Alternative splicing of CEACAM1 by hypoxia-inducible factor-1α enhances tolerance to hepatic ischemia in mice and humans
- PMID: 37531413
- PMCID: PMC11164245
- DOI: 10.1126/scitranslmed.adf2059
Alternative splicing of CEACAM1 by hypoxia-inducible factor-1α enhances tolerance to hepatic ischemia in mice and humans
Abstract
Although alternative splicing (AS) drives transcriptional responses and cellular adaptation to environmental stresses, its contributions in organ transplantation have not been appreciated. We have shown that carcinoembryonic antigen-related cell adhesion molecule (Ceacam1; CD66a), a transmembrane biliary glycoprotein expressed in epithelial, endothelial, and immune cells, determines donor liver transplant quality. Here, we studied how AS of Ceacam1 affects ischemia-reperfusion injury (IRI) in mouse and human livers. We found that the short cytoplasmic isoform Ceacam1-S increased during early acute and late resolution phases of warm IRI injury in mice. Transfection of Ceacam1-deficient mouse hepatocytes with adenoviral Ceacam1-S mitigated hypoxia-induced loss of cellular adhesion by repressing the Ask1/p-p38 cell death pathway. Nucleic acid-blocking morpholinos, designed to selectively induce Ceacam1-S, protected hepatocyte cultures against temperature-induced stress in vitro. Luciferase and chromatin immunoprecipitation assays identified direct binding of hypoxia-inducible factor-1α (Hif-1α) to the mouse polypyrimidine tract binding protein 1 (Ptbp1) promoter region. Dimethyloxalylglycine protected mouse livers from warm IR stress and hepatocellular damage by inhibiting prolyl hydroxylase domain-containing protein 1 and promoting AS of Ceacam1-S. Last, analysis of 46 human donor liver grafts revealed that CEACAM1-S positively correlated with pretransplant HIF1A expression. This also correlated with better transplant outcomes, including reduced TIMP1, total bilirubin, proinflammatory MCP1, CXCL10 cytokines, immune activation markers IL17A, and incidence of delayed complications from biliary anastomosis. This translational study identified mouse Hif-1α-controlled AS of Ceacam1, through transcriptional regulation of Ptbp1 promoter region, as a functional underpinning of hepatoprotection against IR stress and tissue damage in liver transplantation.
Conflict of interest statement
Figures
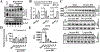




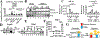


References
-
- Han SP, Tang YH, Smith R, Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430, 379–392 (2010). - PubMed
-
- Wong RJ, Singal AK, Trends in Liver Disease Etiology Among Adults Awaiting Liver Transplantation in the United States, 2014–2019. JAMA Netw Open 3, e1920294 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

