SUMO-activated target traps (SATTs) enable the identification of a comprehensive E3-specific SUMO proteome
- PMID: 37531430
- PMCID: PMC10396300
- DOI: 10.1126/sciadv.adh2073
SUMO-activated target traps (SATTs) enable the identification of a comprehensive E3-specific SUMO proteome
Abstract
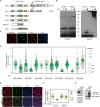
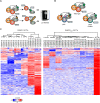
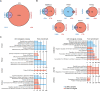
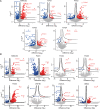
Ubiquitin and ubiquitin-like conjugation cascades consist of dedicated E1, E2, and E3 enzymes with E3s providing substrate specificity. Mass spectrometry-based approaches have enabled the identification of more than 6500 SUMO2/3 target proteins. The limited number of SUMO E3s provides the unique opportunity to systematically study E3 substrate wiring. We developed SUMO-activated target traps (SATTs) and systematically identified substrates for eight different SUMO E3s, PIAS1, PIAS2, PIAS3, PIAS4, NSMCE2, ZNF451, LAZSUL (ZNF451-3), and ZMIZ2. SATTs enabled us to identify 427 SUMO1 and 961 SUMO2/3 targets in an E3-specific manner. We found pronounced E3 substrate preference. Quantitative proteomics enabled us to measure substrate specificity of E3s, quantified using the SATT index. Furthermore, we developed the Polar SATTs web-based tool to browse the dataset in an interactive manner. Overall, we uncover E3-to-target wiring of 1388 SUMO substrates, highlighting unique and overlapping sets of substrates for eight different SUMO E3 ligases.
Figures


References
-
- F. Trulsson, A. C. O. Vertegaal, Site-specific proteomic strategies to identify ubiquitin and SUMO modifications: Challenges and opportunities. Semin Cell Dev. Biol. 132, 97–108 (2022). - PubMed
-
- V. Akimov, I. Barrio-Hernandez, S. V. F. Hansen, P. Hallenborg, A.-K. Pedersen, D. B. Bekker-Jensen, M. Puglia, S. D. K. Christensen, J. T. Vanselow, M. M. Nielsen, I. Kratchmarova, C. D. Kelstrup, J. V. Olsen, B. Blagoev, UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

