Development of an Affinity-Based Probe to Profile Endogenous Human Adenosine A3 Receptor Expression
- PMID: 37531576
- PMCID: PMC10461224
- DOI: 10.1021/acs.jmedchem.3c00854
Development of an Affinity-Based Probe to Profile Endogenous Human Adenosine A3 Receptor Expression
Abstract
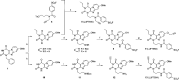
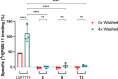
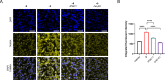
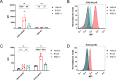
The adenosine A3 receptor (A3AR) is a G protein-coupled receptor (GPCR) that exerts immunomodulatory effects in pathophysiological conditions such as inflammation and cancer. Thus far, studies toward the downstream effects of A3AR activation have yielded contradictory results, thereby motivating the need for further investigations. Various chemical and biological tools have been developed for this purpose, ranging from fluorescent ligands to antibodies. Nevertheless, these probes are limited by their reversible mode of binding, relatively large size, and often low specificity. Therefore, in this work, we have developed a clickable and covalent affinity-based probe (AfBP) to target the human A3AR. Herein, we show validation of the synthesized AfBP in radioligand displacement, SDS-PAGE, and confocal microscopy experiments as well as utilization of the AfBP for the detection of endogenous A3AR expression in flow cytometry experiments. Ultimately, this AfBP will aid future studies toward the expression and function of the A3AR in pathologies.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

