Sézary syndrome originates from heavily mutated hematopoietic progenitors
- PMID: 37531660
- PMCID: PMC10514084
- DOI: 10.1182/bloodadvances.2022008562
Sézary syndrome originates from heavily mutated hematopoietic progenitors
Abstract
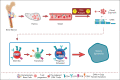
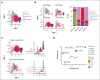
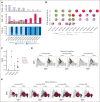
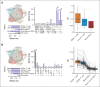
The pathogenesis of cutaneous T-cell lymphoma (CTCL) remains unclear. Using single-cell RNA or T-cell receptor (TCR) sequencing of 32 619 CD3+CD4+ and CD26+/CD7+ and 29 932 CD3+CD4+ and CD26-/CD7- lymphocytes from the peripheral blood of 7 patients with CTCL, coupled to single-cell ATAC-sequencing of 26,411 CD3+CD4+ and CD26+/CD7+ and 33 841 CD3+CD4+ and CD26-/CD7- lymphocytes, we show that tumor cells in Sézary syndrome and mycosis fungoides (MF) exhibit different phenotypes and trajectories of differentiation. When compared to MF, Sézary cells exhibit narrower repertoires of TCRs and exhibit clonal enrichment. Surprisingly, we identified ≥200 mutations in hematopoietic stem cells from multiple patients with Sézary syndrome. Mutations in key oncogenes were also present in peripheral Sézary cells, which also showed the hallmarks of recent thymic egression. Together our data suggest that CTCL arises from mutated lymphocyte progenitors that acquire TCRs in the thymus, which complete their malignant transformation in the periphery.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: J.P.-I. reports funding from TG Therapeutics, MEI, and Viracta, and consulting fees from Janssen, AbbVie, TG Therapeutics, Novartis, Takeda, and AstraZeneca. L.S. reports consulting fees from Dren Bio Inc; is an adviser for Kyowa-Kirin, Inc, Daiichi-Sankyo, and Kymera Therapeutics; and reports clinical research funding from Kyowa Kirin and EUSA Pharma, LLC. J.R.C.-G. reports funding and consulting fees from Anixa Biosciences; consulting fees from Alloy Therapeutics; and stock options from Compass Therapeutics, Anixa Biosciences, and Alloy Therapeutics. The remaining authors declare no competing financial interests.
Figures





References
-
- Beltran BE, Castro D, De La Cruz-Vargas JA, et al. The neutrophil-lymphocyte ratio is prognostic in patients with early stage aggressive peripheral T cell lymphoma. Br J Haematol. 2019;184(4):650–653. - PubMed
-
- Rodd AL, Ververis K, Karagiannis TC. Current and emerging therapeutics for cutaneous T-cell lymphoma: histone deacetylase inhibitors. Lymphoma. 2012;2012:1–10.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

