Development of Plasmodium falciparum liver-stages in hepatocytes derived from human fetal liver organoid cultures
- PMID: 37532704
- PMCID: PMC10397232
- DOI: 10.1038/s41467-023-40298-7
Development of Plasmodium falciparum liver-stages in hepatocytes derived from human fetal liver organoid cultures
Abstract
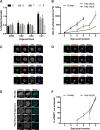
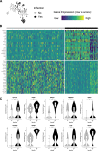
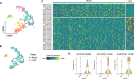
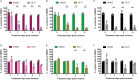
Plasmodium falciparum (Pf) parasite development in liver represents the initial step of the life-cycle in the human host after a Pf-infected mosquito bite. While an attractive stage for life-cycle interruption, understanding of parasite-hepatocyte interaction is inadequate due to limitations of existing in vitro models. We explore the suitability of hepatocyte organoids (HepOrgs) for Pf-development and show that these cells permitted parasite invasion, differentiation and maturation of different Pf strains. Single-cell messenger RNA sequencing (scRNAseq) of Pf-infected HepOrg cells has identified 80 Pf-transcripts upregulated on day 5 post-infection. Transcriptional profile changes are found involving distinct metabolic pathways in hepatocytes with Scavenger Receptor B1 (SR-B1) transcripts highly upregulated. A novel functional involvement in schizont maturation is confirmed in fresh primary hepatocytes. Thus, HepOrgs provide a strong foundation for a versatile in vitro model for Pf liver-stages accommodating basic biological studies and accelerated clinical development of novel tools for malaria control.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- WHO. World Malaria Report 2021 (WHO, 2021).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

