Africa-specific human genetic variation near CHD1L associates with HIV-1 load
- PMID: 37532928
- PMCID: PMC10848312
- DOI: 10.1038/s41586-023-06370-4
Africa-specific human genetic variation near CHD1L associates with HIV-1 load
Erratum in
-
Author Correction: Africa-specific human genetic variation near CHD1L associates with HIV-1 load.Nature. 2023 Sep;621(7979):E42. doi: 10.1038/s41586-023-06591-7. Nature. 2023. PMID: 37670157 No abstract available.
Abstract
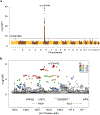
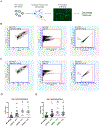
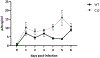
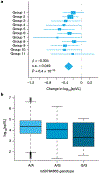
HIV-1 remains a global health crisis1, highlighting the need to identify new targets for therapies. Here, given the disproportionate HIV-1 burden and marked human genome diversity in Africa2, we assessed the genetic determinants of control of set-point viral load in 3,879 people of African ancestries living with HIV-1 participating in the international collaboration for the genomics of HIV3. We identify a previously undescribed association signal on chromosome 1 where the peak variant associates with an approximately 0.3 log10-transformed copies per ml lower set-point viral load per minor allele copy and is specific to populations of African descent. The top associated variant is intergenic and lies between a long intergenic non-coding RNA (LINC00624) and the coding gene CHD1L, which encodes a helicase that is involved in DNA repair4. Infection assays in iPS cell-derived macrophages and other immortalized cell lines showed increased HIV-1 replication in CHD1L-knockdown and CHD1L-knockout cells. We provide evidence from population genetic studies that Africa-specific genetic variation near CHD1L associates with HIV replication in vivo. Although experimental studies suggest that CHD1L is able to limit HIV infection in some cell types in vitro, further investigation is required to understand the mechanisms underlying our observations, including any potential indirect effects of CHD1L on HIV spread in vivo that our cell-based assays cannot recapitulate.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
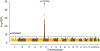

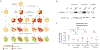
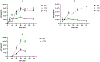
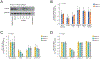
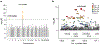
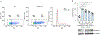
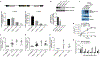
References
-
- UNAIDS Data 2021 (UNAIDS, 2021); https://www.unaids.org/en/resources/documents/2021/2021_unaids_data.
-
- Gurdasani D, Barroso I, Zeggini E & Sandhu MS Genomics of disease risk in globally diverse populations. Nat. Rev. Genet 20, 520–535 (2019). - PubMed
-
- Prevention Gap Report (UNAIDS, 2016).
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_00033/3/MRC_/Medical Research Council/United Kingdom
- 75N91019D00024/CA/NCI NIH HHS/United States
- U01 HL146208/HL/NHLBI NIH HHS/United States
- MR/N02043X/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- U01 HL146240/HL/NHLBI NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- U24 HG006941/HG/NHGRI NIH HHS/United States
- U01 HL146333/HL/NHLBI NIH HHS/United States
- R01 AI165236/AI/NIAID NIH HHS/United States
- MR/S009752/1/MRC_/Medical Research Council/United Kingdom
- UM1 AI068634/AI/NIAID NIH HHS/United States
- MC_UU_00033/1/MRC_/Medical Research Council/United Kingdom
- U01 HL146201/HL/NHLBI NIH HHS/United States
- P30 AI117943/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- U54 AI170792/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

