Synergy and oxygen adaptation for development of next-generation probiotics
- PMID: 37532933
- PMCID: PMC10412450
- DOI: 10.1038/s41586-023-06378-w
Synergy and oxygen adaptation for development of next-generation probiotics
Abstract
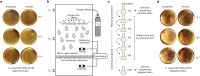
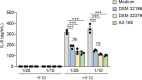
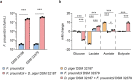
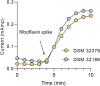
The human gut microbiota has gained interest as an environmental factor that may contribute to health or disease1. The development of next-generation probiotics is a promising strategy to modulate the gut microbiota and improve human health; however, several key candidate next-generation probiotics are strictly anaerobic2 and may require synergy with other bacteria for optimal growth. Faecalibacterium prausnitzii is a highly prevalent and abundant human gut bacterium associated with human health, but it has not yet been developed into probiotic formulations2. Here we describe the co-isolation of F. prausnitzii and Desulfovibrio piger, a sulfate-reducing bacterium, and their cross-feeding for growth and butyrate production. To produce a next-generation probiotic formulation, we adapted F. prausnitzii to tolerate oxygen exposure, and, in proof-of-concept studies, we demonstrate that the symbiotic product is tolerated by mice and humans (ClinicalTrials.gov identifier: NCT03728868 ) and is detected in the human gut in a subset of study participants. Our study describes a technology for the production of next-generation probiotics based on the adaptation of strictly anaerobic bacteria to tolerate oxygen exposures without a reduction in potential beneficial properties. Our technology may be used for the development of other strictly anaerobic strains as next-generation probiotics.
© 2023. The Author(s).
Conflict of interest statement
M.T.K. is employed in part by Metabogen AB and F.B. is founder of Metabogen AB. Metabogen AB has contributed with economic support and product for the human intervention, but was not involved in analyses of the data. F.B. receives research funding from Biogaia AB and is on the scientific advisory board of Bactolife A/S. The other authors declare no competing interests.
Figures






Comment in
-
Turning cooperative bacteria into probiotics for human health.Nature. 2023 Aug;620(7973):283-284. doi: 10.1038/d41586-023-02407-w. Nature. 2023. PMID: 37532854 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources
Medical

