Noncoding RNAs in apoptosis: identification and function
- PMID: 37533667
- PMCID: PMC10393110
- DOI: 10.3906/biy-2109-35
Noncoding RNAs in apoptosis: identification and function
Abstract
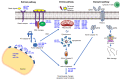
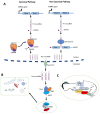
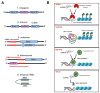
Apoptosis is a vital cellular process that is critical for the maintenance of homeostasis in health and disease. The derailment of apoptotic mechanisms has severe consequences such as abnormal development, cancer, and neurodegenerative diseases. Thus, there exist complex regulatory mechanisms in eukaryotes to preserve the balance between cell growth and cell death. Initially, protein-coding genes were prioritized in the search for such regulatory macromolecules involved in the regulation of apoptosis. However, recent genome annotations and transcriptomics studies have uncovered a plethora of regulatory noncoding RNAs that have the ability to modulate not only apoptosis but also many other biochemical processes in eukaryotes. In this review article, we will cover a brief summary of apoptosis and detection methods followed by an extensive discussion on microRNAs, circular RNAs, and long noncoding RNAs in apoptosis.
Keywords: Apoptosis; circular RNA; long noncoding RNA; microRNA; noncoding RNA.
© TÜBİTAK.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interests.
Figures

References
-
- Alkan AH, Akgül B. Endogenous miRNA sponges. Methods in Molecular Biology. 2022;2257:91–104. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
