Transcriptomic profiles of multiple organ dysfunction syndrome phenotypes in pediatric critical influenza
- PMID: 37533854
- PMCID: PMC10390830
- DOI: 10.3389/fimmu.2023.1220028
Transcriptomic profiles of multiple organ dysfunction syndrome phenotypes in pediatric critical influenza
Abstract
Background: Influenza virus is responsible for a large global burden of disease, especially in children. Multiple Organ Dysfunction Syndrome (MODS) is a life-threatening and fatal complication of severe influenza infection.
Methods: We measured RNA expression of 469 biologically plausible candidate genes in children admitted to North American pediatric intensive care units with severe influenza virus infection with and without MODS. Whole blood samples from 191 influenza-infected children (median age 6.4 years, IQR: 2.2, 11) were collected a median of 27 hours following admission; for 45 children a second blood sample was collected approximately seven days later. Extracted RNA was hybridized to NanoString mRNA probes, counts normalized, and analyzed using linear models controlling for age and bacterial co-infections (FDR q<0.05).
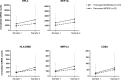
Results: Comparing pediatric samples collected near admission, children with Prolonged MODS for ≥7 days (n=38; 9 deaths) had significant upregulation of nine mRNA transcripts associated with neutrophil degranulation (RETN, TCN1, OLFM4, MMP8, LCN2, BPI, LTF, S100A12, GUSB) compared to those who recovered more rapidly from MODS (n=27). These neutrophil transcripts present in early samples predicted Prolonged MODS or death when compared to patients who recovered, however in paired longitudinal samples, they were not differentially expressed over time. Instead, five genes involved in protein metabolism and/or adaptive immunity signaling pathways (RPL3, MRPL3, HLA-DMB, EEF1G, CD8A) were associated with MODS recovery within a week.
Conclusion: Thus, early increased expression of neutrophil degranulation genes indicated worse clinical outcomes in children with influenza infection, consistent with reports in adult cohorts with influenza, sepsis, and acute respiratory distress syndrome.
Keywords: MODS; critical care; influenza; neutrophil degranulation; neutrophil transcripts; organ failure; pediatric intensive care; sepsis.
Copyright © 2023 Novak, Crawford, Hahn, Hall, Thair, Newhams, Chou, Mourani, Tarquinio, Markovitz, Loftis, Weiss, Higgerson, Schwarz, Pinto, Thomas, Gedeit, Sanders, Mahapatra, Coates, Cvijanovich, Ackerman, Tellez, McQuillen, Kurachek, Shein, Lange, Thomas and Randolph.
Conflict of interest statement
AGR, TN, and MMN: NIH/NIAID as declared in the funding statement - grant support paid to Boston Children’s Hospital. Also current CDC grant for COVID-19 work unrelated to current manuscript also paid to Boston Children’s Hospital under PI AGR. AGR also receives Royalties from UpToDate, Inc. as Section editor of Pediatric Critical Care Medicine. Honoraria: Grand Rounds presentations on MIS-C not related to current manuscript. Participation on a Data Safety Monitoring/Advisory Board: NIH Grace Study. Leadership or fiduciary role in other board, society, committee or advocacy group, paid or unpaid: Medical Advisory Board for Families Fight Flu and Chair of International Sepsis Forum. JCC: additionally declares support from ALSAC for the current work. Other entities not related to the current work: U01AI150747, R01AI136514, U01AI144616; American Association of Immunologists Abstract award 2021, Patent: Methods for Treating or Reducing the Severity of a Viral Infection Publication no. WO 2021/226174 A1. MWH declared Kiadis, Licensing income, unrelated to the content of this manuscript, American Board of Pediatrics Payment, Abbive payment. Partner Therapeutics Provision of study drug, unrelated to the current manuscript, Sobi Provision of study drug, unrelated to the current manuscript. JC: NIH, paid to institution not related to this work; GLG Group payment to her, Elsevier payment made to her for work done as Associate Editor; Patent pending for Methods and compositions for treating and preventing T cell-driven diseases. BMC: Not related to current work: receives grants paid to institution from American Lung Association. Sobi Participation on a Data Safety Monitoring Board/Advisory Board. SLW: Receives royalties from UpToDate, Inc. Has grants/contracts with Pennsylvania Dept of Health not related to current manuscript. PGT: declares support from ALSAC and NIAID R01 AI154470 for the current work. Has several grants not associated with current work: NIAID 5R01AI128805-05; 1R01AI154470; NIAID 75N93019C00052; 5R01AI136514; 5R01AI35025; NIAID 5U01AI144616; NIAID U01AI150747. Royalties/licenses paid to both PGT and his institution: TCR cloning technology—Miltenyi Biotec & TCR amplification technology—Shennon Bio. Consulting fees paid to him: Johnson and Johnson, Cytoagents, Immunoscape, Shennon Bio. Payment or honoraria for lectures, presentations, or other events: Illumina—Future Genomic Advances, Yale University, CZ Biohub, PACT Pharma, UCSD, Tufts, University of Arizona, Mt. Sinai, Umass, OSU, Korean Association of Immunology, SISMID, University of Washington, MSKCC, Washington University, University of Missouri, Fred Hutch, UNM Illumina Single Cell, Cincinnati Childrens, University of Toronto, Purdue University, Iowa State University, University of Georgia. Patent: Methods for Treating or Reducing the Severity of a Viral Infection Publication no. WO 2021/226174 A1. (continued disclosures for PGT:) Support for attending meetings and/or travel: NIH Study Section, Keystone Viral Immunity, NIAID CEIRR, CEIRS and CIVIC meetings, GRC, 10X Users Symposium, Illumina Symposia, ImmunOktoberfest, Options for the Control of Influenza XI, Carghese Workshop, FOCIS, Max Planck for Complex Systems, Santa Fe Institute, AAI, Weizmann Institute, APS Society, Banff Institute, ISIRV meeting, University of Melbourne, British Society of Immunology. Stock or stock options: Shennon Bio Scientific advisory board payments. (Additional patents:) US20170304293A1 Coordinated metabolic reprogramming in response to productive viral infections; Cloning and expression system for t-cell receptors; License payments made to me and institution; Method for detecting SARS-CoV-2 infection. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- FluSurv-NET influenza hospitalization surveillance network. Laboratory-Confirmed Influenza Hospitalizations: Centers for Disease Control and Prevention; (2022) Atlanta, GA, United States: FluSurv-NET: Influenza Hospitalization Surveillance Network. Available at: https://gis.cdc.gov/grasp/fluview/fluhosprates.html.
-
- FluSurv-NET: influenza-associated pediatric mortality surveillance network. Influenza Associated Pediatric Deaths: Centers for Disease Control and Prevention; (2022). Atlanta, GA, United States: Available at: https://gis.cdc.gov/GRASP/Fluview/PedFluDeath.html.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

