Epigenetic regulation of nuclear processes in fungal plant pathogens
- PMID: 37535497
- PMCID: PMC10399791
- DOI: 10.1371/journal.ppat.1011525
Epigenetic regulation of nuclear processes in fungal plant pathogens
Abstract
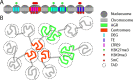
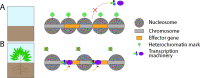
Through the association of protein complexes to DNA, the eukaryotic nuclear genome is broadly organized into open euchromatin that is accessible for enzymes acting on DNA and condensed heterochromatin that is inaccessible. Chemical and physical alterations to chromatin may impact its organization and functionality and are therefore important regulators of nuclear processes. Studies in various fungal plant pathogens have uncovered an association between chromatin organization and expression of in planta-induced genes that are important for pathogenicity. This review discusses chromatin-based regulation mechanisms as determined in the fungal plant pathogen Verticillium dahliae and relates the importance of epigenetic transcriptional regulation and other nuclear processes more broadly in fungal plant pathogens.
Copyright: © 2023 Kramer et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections.New Phytol. 2017 Jul;215(1):368-381. doi: 10.1111/nph.14537. Epub 2017 Apr 13. New Phytol. 2017. PMID: 28407259
-
A fungal milRNA mediates epigenetic repression of a virulence gene in Verticillium dahliae.Philos Trans R Soc Lond B Biol Sci. 2019 Mar 4;374(1767):20180309. doi: 10.1098/rstb.2018.0309. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30967013 Free PMC article.
-
Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens.PLoS Pathog. 2011 Jul;7(7):e1002137. doi: 10.1371/journal.ppat.1002137. Epub 2011 Jul 28. PLoS Pathog. 2011. PMID: 21829347 Free PMC article.
-
Epigenetic regulation of development and pathogenesis in fungal plant pathogens.Mol Plant Pathol. 2017 Aug;18(6):887-898. doi: 10.1111/mpp.12499. Epub 2016 Nov 28. Mol Plant Pathol. 2017. PMID: 27749982 Free PMC article. Review.
-
Nuclear architecture and gene regulation.Biochim Biophys Acta. 2008 Nov;1783(11):2174-84. doi: 10.1016/j.bbamcr.2008.07.018. Epub 2008 Jul 31. Biochim Biophys Acta. 2008. PMID: 18718493 Review.
Cited by
-
Starship giant transposons dominate plastic genomic regions in a fungal plant pathogen and drive virulence evolution.Nat Commun. 2025 Jul 24;16(1):6806. doi: 10.1038/s41467-025-61986-6. Nat Commun. 2025. PMID: 40707455 Free PMC article.
-
Analysis of histone modification interplay reveals two distinct domains in facultative heterochromatin in Pyricularia oryzae.Commun Biol. 2025 Jul 22;8(1):1086. doi: 10.1038/s42003-025-08473-2. Commun Biol. 2025. PMID: 40695975 Free PMC article.
-
Dimensions of genome dynamics in fungal pathogens: from fundamentals to applications.BMC Biol. 2024 Jan 26;22(1):19. doi: 10.1186/s12915-023-01786-w. BMC Biol. 2024. PMID: 38279095 Free PMC article.
-
RNAi epimutations conferring antifungal drug resistance are inheritable.Nat Commun. 2025 Aug 7;16(1):7293. doi: 10.1038/s41467-025-62572-6. Nat Commun. 2025. PMID: 40775213 Free PMC article.
-
Epigenetic modulation of fungal pathogens: a focus on Magnaporthe oryzae.Front Microbiol. 2024 Oct 28;15:1463987. doi: 10.3389/fmicb.2024.1463987. eCollection 2024. Front Microbiol. 2024. PMID: 39529673 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

