Understanding the efficacy of wastewater surveillance for SARS-CoV-2 in two diverse communities
- PMID: 37535602
- PMCID: PMC10399835
- DOI: 10.1371/journal.pone.0289343
Understanding the efficacy of wastewater surveillance for SARS-CoV-2 in two diverse communities
Abstract
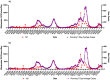
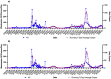
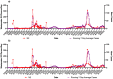
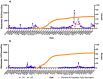
During the COVID-19 pandemic, wastewater-based surveillance has been shown to be a useful tool for monitoring the spread of disease in communities and the emergence of new viral variants of concern. As the pandemic enters its fourth year and clinical testing has declined, wastewater offers a consistent non-intrusive way to monitor community health in the long term. This study sought to understand how accurately wastewater monitoring represented the actual burden of disease between communities. Two communities varying in size and demographics in Michigan were monitored for SARS-CoV-2 in wastewater between March of 2020 and February of 2022. Additionally, each community was monitored for SARS-CoV-2 variants of concern from December 2020 to February 2022. Wastewater results were compared with zipcode and county level COVID-19 case data to determine which scope of clinical surveillance was most correlated with wastewater loading. Pearson r correlations were highest in the smaller of the two communities (population of 25,000) for N1 GC/person/day with zipcode level case data, and date of the onset of symptoms (r = 0.81). A clear difference was seen with more cases and virus signals in the wastewater of the larger community (population 110,000) when examined based on vaccine status, which reached only 50%. While wastewater levels of SARS-CoV-2 had a lower correlation to cases in the larger community, the information was still seen as valuable in supporting public health actions and further data including vaccination status should be examined in the future.
Copyright: © 2023 Flood et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- National Academies of Sciences, Engineering, and Medicine. 2023. Wastewater-based Disease Surveillance for Public Health Action. Washington, DC: The National Academies Press. - PubMed
-
- Ferraro GB; Veneri C; Mancini P; Iaconelli M; Suffredini E; Bonadonna L; et al. A State-of-the-Art Scoping Review on SARS-CoV-2 in Sewage Focusing on the Potential of Wastewater Surveillance for the Monitoring of the COVID-19 Pandemic 2022. Food and Environmental Virology Volume14 Issue4, Page 315–354. doi: 10.1007/s12560-021-09498-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

