Transcriptional signatures measured in whole blood correlate with protection against tuberculosis in inbred and outbred mice
- PMID: 37535648
- PMCID: PMC10399789
- DOI: 10.1371/journal.pone.0289358
Transcriptional signatures measured in whole blood correlate with protection against tuberculosis in inbred and outbred mice
Abstract
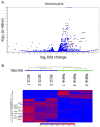
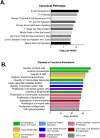
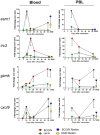
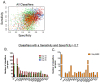
Although BCG has been used for almost 100 years to immunize against Mycobacterium tuberculosis, TB remains a global public health threat. Numerous clinical trials are underway studying novel vaccine candidates and strategies to improve or replace BCG, but vaccine development still lacks a well-defined set of immune correlates to predict vaccine-induced protection against tuberculosis. This study aimed to address this gap by examining transcriptional responses to BCG vaccination in C57BL/6 inbred mice, coupled with protection studies using Diversity Outbred mice. We evaluated relative gene expression in blood obtained from vaccinated mice, because blood is easily accessible, and data can be translated to human studies. We first determined that the average peak time after vaccination is 14 days for gene expression of a small subset of immune-related genes in inbred mice. We then performed global transcriptomic analyses using whole blood samples obtained two weeks after mice were vaccinated with BCG. Using comparative bioinformatic analyses and qRT-PCR validation, we developed a working correlate panel of 18 genes that were highly correlated with administration of BCG but not heat-killed BCG. We then tested this gene panel using BCG-vaccinated Diversity Outbred mice and revealed associations between the expression of a subset of genes and disease outcomes after aerosol challenge with M. tuberculosis. These data therefore demonstrate that blood-based transcriptional immune correlates measured within a few weeks after vaccination can be derived to predict protection against M. tuberculosis, even in outbred populations.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- World Health Organization. Global tuberculosis report 2022. 2022.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

