Comparison of DNA concentration and bacterial pathogen PCR detection when using two DNA extraction kits for nasopharyngeal/oropharyngeal samples
- PMID: 37535692
- PMCID: PMC10399880
- DOI: 10.1371/journal.pone.0289557
Comparison of DNA concentration and bacterial pathogen PCR detection when using two DNA extraction kits for nasopharyngeal/oropharyngeal samples
Abstract
Introduction: Several important human pathogens that cause life-threatening infections are asymptomatically carried in the Nasopharynx/Oropharynx (NP/OP). DNA extraction is a prerequisite for most culture-independent techniques used to identify pathogens in the NP/OP. However, components of DNA extraction kits differ thereby giving rise to differences in performance. We compared the DNA concentration and the detection of three pathogens in the NP/OP using the discontinued DNeasy PowerSoil Kit (Kit DP) and the DNeasy PowerLyzer PowerSoil Kit (Kit DPP).
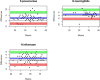
Methods: DNA was extracted from the same set of 103 NP/OP samples using the two kits. DNA concentration was measured using the Qubit 2.0 Fluorometer. Real-time Polymerase Chain reaction (RT-PCR) was done using the QuantStudio 7-flex system to detect three pathogens: S. pneumoniae, H. influenzae, and N. meningitidis. Bland-Altman statistics and plots were used to determine the threshold cycle (Ct) value agreement for the two kits.
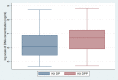
Results: The average DNA concentration from kit DPP was higher than Kit DP; 1235.6 ng/ml (SD = 1368.3) vs 884.9 ng/ml (SD = 1095.3), p = 0.002. Using a Ct value cutoff of 40 for positivity, the concordance for the presence of S. pneumoniae was 82% (84/102); 94%(96/103) for N. meningitidis and 92%(95/103) for H. influenzae. Kit DP proportionately resulted in higher Ct values than Kit DPP for all pathogens. The Ct value bias of measurement for S. pneumoniae was +2.4 (95% CI, 1.9-3.0), +1.4 (95% CI, 0.9-1.9) for N. meningitidis and +1.4 (95% CI, 0.2-2.5) for H. influenzae.
Conclusion: The higher DNA concentration obtained using kit DPP could increase the chances of recovering low abundant bacteria. The PCR results were reproducible for more than 90% of the samples for the gram-negative H. influenzae and N. meningitidis. Ct value variations of the kits must be taken into consideration when comparing studies that have used the two kits.
Copyright: © 2023 Khan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

