Exploring the G-quadruplex binding and unwinding activity of the bacterial FeS helicase DinG
- PMID: 37537265
- PMCID: PMC10400533
- DOI: 10.1038/s41598-023-39675-5
Exploring the G-quadruplex binding and unwinding activity of the bacterial FeS helicase DinG
Abstract
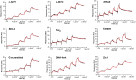
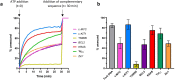
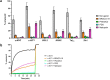
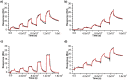
Despite numerous reports on the interactions of G-quadruplexes (G4s) with helicases, systematic analysis addressing the selectivity and specificity of each helicase towards a variety of G4 topologies are scarce. Among the helicases able to unwind G4s are those containing an iron-sulphur (FeS) cluster, including both the bacterial DinG (found in E. coli and several pathogenic bacteria) and the medically important eukaryotic homologues (XPD, FancJ, DDX11 and RTEL1). We carried out a detailed study of the interactions between the E. coli DinG and a variety of G4s, by employing physicochemical and biochemical methodologies. A series of G4-rich sequences from different genomic locations (promoter and telomeric regions), able to form unimolecular G4 structures with diverse topologies, were analyzed (c-KIT1, KRAS, c-MYC, BCL2, Tel23, T30695, Zic1). DinG binds to most of the investigated G4s with little discrimination, while it exhibits a clear degree of unwinding specificity towards different G4 topologies. Whereas previous reports suggested that DinG was active only on bimolecular G4s, here we show that it is also able to bind to and resolve the more physiologically relevant unimolecular G4s. In addition, when the G4 structures were stabilized by ligands (Pyridostatin, PhenDC3, BRACO-19 or Netropsin), the DinG unwinding activity decreased and in most cases was abolished, with a pattern that is not simply explained by a change in binding affinity. Overall, these results have important implications for the biochemistry of helicases, strongly suggesting that when analysing the G4 unwinding property of an enzyme, it is necessary to investigate a variety of G4 substrates.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

