Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization
- PMID: 37537642
- PMCID: PMC10520629
- DOI: 10.1002/advs.202207672
Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization
Abstract
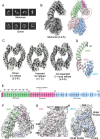
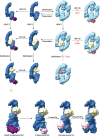
HACE1 is an ankyrin repeat (AKR) containing HECT-type E3 ubiquitin ligase that interacts with and ubiquitinates multiple substrates. While HACE1 is a well-known tumor suppressor, its structure and mode of ubiquitination are not understood. The authors present the cryo-EM structures of human HACE1 along with in vitro functional studies that provide insights into how the enzymatic activity of HACE1 is regulated. HACE1 comprises of an N-terminal AKR domain, a middle (MID) domain, and a C-terminal HECT domain. Its unique G-shaped architecture interacts as a homodimer, with monomers arranged in an antiparallel manner. In this dimeric arrangement, HACE1 ubiquitination activity is hampered, as the N-terminal helix of one monomer restricts access to the C-terminal domain of the other. The in vitro ubiquitination assays, hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis, mutagenesis, and in silico modeling suggest that the HACE1 MID domain plays a crucial role along with the AKRs in RAC1 substrate recognition.
Keywords: HACE1; HECT E3 ligases; RAC1; cancer; ubiquitination.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Komander D., Rape M., Annu. Rev. Biochem. 2012, 81, 203. - PubMed
-
- Ulrich H. D., Walden H., Nat. Rev. Mol. Cell Biol. 2010, 11, 479. - PubMed
-
- Kerscher O., Felberbaum R., Hochstrasser M., Annu. Rev. Cell Dev. Biol. 2006, 22, 159. - PubMed
-
- Singh S., Ng J., Sivaraman J., Pharmacol. Ther. 2021, 224, 107809. - PubMed
-
- Scheffner M., Nuber U., Huibregtse J. M., Nature 1995, 373, 81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
