Members of the CUGBP Elav-like family of RNA-binding proteins are expressed in distinct populations of primary sensory neurons
- PMID: 37537886
- PMCID: PMC11792980
- DOI: 10.1002/cne.25520
Members of the CUGBP Elav-like family of RNA-binding proteins are expressed in distinct populations of primary sensory neurons
Abstract
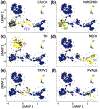
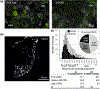
Primary sensory dorsal root ganglia (DRG) neurons are diverse, with distinct populations that respond to specific stimuli. Previously, we observed that functionally distinct populations of DRG neurons express mRNA transcript variants with different 3' untranslated regions (3'UTRs). 3'UTRs harbor binding sites for interaction with RNA-binding proteins (RBPs) for transporting mRNAs to subcellular domains, modulating transcript stability, and regulating the rate of translation. In the current study, analysis of publicly available single-cell RNA-sequencing data generated from adult mice revealed that 17 3'UTR-binding RBPs were enriched in specific populations of DRG neurons. This included four members of the CUG triplet repeat (CUGBP) Elav-like family (CELF): CELF2 and CELF4 were enriched in peptidergic, CELF6 in both peptidergic and nonpeptidergic, and CELF3 in tyrosine hydroxylase-expressing neurons. Immunofluorescence studies confirmed that 60% of CELF4+ neurons are small-diameter C fibers and 33% medium-diameter myelinated (likely Aδ) fibers and showed that CELF4 is distributed to peripheral termini. Coexpression analyses using transcriptomic data and immunofluorescence revealed that CELF4 is enriched in nociceptive neurons that express GFRA3, CGRP, and the capsaicin receptor TRPV1. Reanalysis of published transcriptomic data from macaque DRG revealed a highly similar distribution of CELF members, and reanalysis of single-nucleus RNA-sequencing data derived from mouse and rat DRG after sciatic injury revealed differential expression of CELFs in specific populations of sensory neurons. We propose that CELF RBPs may regulate the fate of mRNAs in populations of nociceptors, and may play a role in pain and/or neuronal regeneration following nerve injury.
Keywords: 3′UTR; CELF; RNA-binding protein; dorsal root ganglion; nociceptor; sensory.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
The authors declare no competing financial interests.
Figures






References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, & Sherlock G (2000). Gene Ontology: Tool for the unification of biology. Nature Genetics, 25(1), Article 1. 10.1038/75556 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

