High-Dimensional MR Spatiospectral Imaging by Integrating Physics-Based Modeling and Data-Driven Machine Learning: Current progress and future directions
- PMID: 37538148
- PMCID: PMC10398845
- DOI: 10.1109/msp.2022.3203867
High-Dimensional MR Spatiospectral Imaging by Integrating Physics-Based Modeling and Data-Driven Machine Learning: Current progress and future directions
Abstract
Magnetic resonance spectroscopic imaging (MRSI) offers a unique molecular window into the physiological and pathological processes in the human body. However, the applications of MRSI have been limited by a number of long-standing technical challenges due to high dimensionality and low signal-to-noise ratio (SNR). Recent technological developments integrating physics-based modeling and data-driven machine learning that exploit unique physical and mathematical properties of MRSI signals have demonstrated impressive performance in addressing these challenges for rapid, high-resolution, quantitative MRSI. This paper provides a systematic review of these progresses in the context of MRSI physics and offers perspectives on promising future directions.
Figures
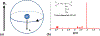
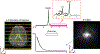
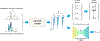
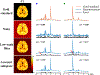
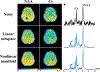

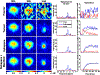
Similar articles
-
High-resolution (1) H-MRSI of the brain using SPICE: Data acquisition and image reconstruction.Magn Reson Med. 2016 Oct;76(4):1059-70. doi: 10.1002/mrm.26019. Epub 2015 Oct 28. Magn Reson Med. 2016. PMID: 26509928 Free PMC article.
-
Constrained Magnetic Resonance Spectroscopic Imaging by Learning Nonlinear Low-Dimensional Models.IEEE Trans Med Imaging. 2020 Mar;39(3):545-555. doi: 10.1109/TMI.2019.2930586. Epub 2019 Jul 23. IEEE Trans Med Imaging. 2020. PMID: 31352337
-
Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces.Magn Reson Med. 2020 Feb;83(2):377-390. doi: 10.1002/mrm.27980. Epub 2019 Sep 4. Magn Reson Med. 2020. PMID: 31483526 Free PMC article.
-
Fast data acquisition techniques in magnetic resonance spectroscopic imaging.NMR Biomed. 2019 Mar;32(3):e4046. doi: 10.1002/nbm.4046. Epub 2019 Jan 14. NMR Biomed. 2019. PMID: 30637822 Review.
-
In vivo MR spectroscopic imaging of the prostate, from application to interpretation.Anal Biochem. 2017 Jul 15;529:158-170. doi: 10.1016/j.ab.2017.02.001. Epub 2017 Feb 3. Anal Biochem. 2017. PMID: 28167072 Review.
Cited by
-
Deep-ER: Deep Learning ECCENTRIC Reconstruction for fast high-resolution neurometabolic imaging.Neuroimage. 2025 Apr 1;309:121045. doi: 10.1016/j.neuroimage.2025.121045. Epub 2025 Feb 1. Neuroimage. 2025. PMID: 39894238 Free PMC article.
-
High-resolution 1H-MRSI at 9.4 T by integrating relaxation enhancement and subspace imaging.NMR Biomed. 2024 Oct;37(10):e5161. doi: 10.1002/nbm.5161. Epub 2024 May 8. NMR Biomed. 2024. PMID: 38715469 Free PMC article.
-
MR Spatiospectral Reconstruction Integrating Subspace Modeling and Self-Supervised Spatiotemporal Denoising.IEEE Trans Med Imaging. 2025 Jul;44(7):3002-3011. doi: 10.1109/TMI.2025.3555928. IEEE Trans Med Imaging. 2025. PMID: 40153290 Free PMC article.
References
-
- Lauterbur PC, Kramer DM, House WV, and Chen C-N, “Zeugmatographic high resolution nuclear magnetic resonance spectroscopy: Images of chemical inhomogeneity within macroscopic objects,” J. Amer. Chem. Soc, vol. 97, pp. 6866–6868, 1975.
-
- Ross B and Michaelis T, “Clinical applications of magnetic resonance spectroscopy.” Magn. Reson. Q, vol. 10, no. 4, pp. 191–247, 1994. - PubMed
-
- de Graaf RA, In Vivo NMR Spectroscopy: Principles and Techniques. Hoboken, NJ: John Wiley & Sons, 2018.
Grants and funding
LinkOut - more resources
Full Text Sources
