The genome sequence of the turban top shell, Gibbula magus (Linnaeus, 1758)
- PMID: 37538380
- PMCID: PMC10394393
- DOI: 10.12688/wellcomeopenres.18792.1
The genome sequence of the turban top shell, Gibbula magus (Linnaeus, 1758)
Abstract
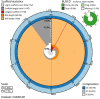
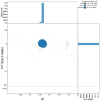
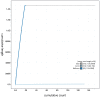
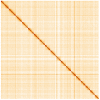
We present a genome assembly from an individual Gibbula magus (the turban top shell; Mollusca; Gastropoda; Trochida; Trochidae). The genome sequence is 1,470 megabases in span. Most of the assembly is scaffolded into 18 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 16.1 kilobases in length. Gene annotation of this assembly on Ensembl identified 41,167 protein coding genes.
Keywords: Gibbula magus; Trochida; chromosomal; genome sequence; turban top shell.
Copyright: © 2023 Adkins P et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
-
- Affenzeller S, Haar N, Steiner G: Revision of the genus complex Gibbula: an integrative approach to delineating the Eastern Mediterranean genera Gibbula Risso, 1826, Steromphala Gray, 1847, and Phorcus Risso, 1826 using DNA-barcoding and geometric morphometrics (Vetigastropoda, Trochoidea). Organisms Diversity & Evolution. 2017;17(4):789–812. 10.1007/s13127-017-0343-5 - DOI
-
- Anistratenko VV: Lectotypes for Tricolia pullus, Gibbula divaricata and Theodoxus fluviatilis (Mollusca, Gastropoda) revisited. Vestnik Zoologii. 2005;39(6):3–10. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources

