Identification of new theobromine-based derivatives as potent VEGFR-2 inhibitors: design, semi-synthesis, biological evaluation, and in silico studies
- PMID: 37538515
- PMCID: PMC10395314
- DOI: 10.1039/d3ra04007k
Identification of new theobromine-based derivatives as potent VEGFR-2 inhibitors: design, semi-synthesis, biological evaluation, and in silico studies
Abstract
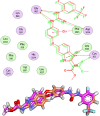
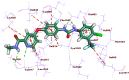
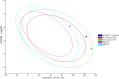
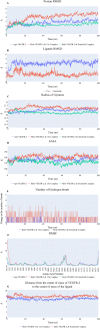
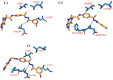
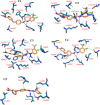
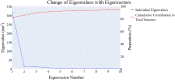
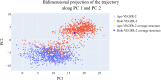
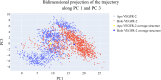
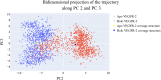
This study aimed to design anticancer theobromine derivatives inhibiting VEGFR-2. The new compounds were tested in vitro to evaluate their effectiveness against MCF-7 and HepG2 cancer cell lines. Among these compounds, 15a showed the highest cytotoxicity against HepG2, with an IC50 value of 0.76 μM, and significant anti-proliferative effects on MCF-7, with an IC50 value of 1.08 μM. Notably, the selectivity index of 15a against the two cancer cells was 98.97 and 69.64, respectively. Moreover, 15a demonstrated potent VEGFR-2 inhibitory activity (IC50 = 0.239 μM). Further investigations revealed that 15a induced apoptosis in HepG2 cells, significantly increasing early-stage and late-stage apoptosis percentages from 3.06% and 0.71% to 29.49% and 9.63%, respectively. It also upregulated caspase-3 and caspase-9 levels by 3.45-fold and 2.37-fold, respectively compared to control HepG2 cells. Additionally, 15a inhibited the migration and wound healing ability of HepG2 cells. Molecular docking confirmed the binding affinities of the semi-synthesized compounds to VEGFR-2, consistent with in vitro results. Several computational analyses (DFT, MD simulations, MM-GBSA, PLIP, and essential dynamics) supported the stability of the 15a-VEGFR-2 complex. Overall, the biological and computational findings suggest that compound 15a could be a promising lead compound for the development of a novel apoptotic anticancer agent.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures











References
-
- Bray F. Ferlay J. Soerjomataram I. Siegel R. L. Torre L. A. Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Belal A. Abdel Gawad N. M. Mehany A. B. Abourehab M. A. Elkady H. Al-Karmalawy A. A. Ismael A. S. Design, synthesis and molecular docking of new fused 1 H-pyrroles, pyrrolo [3,2-d] pyrimidines and pyrrolo [3,2-e][1,4] diazepine derivatives as potent EGFR/CDK2 inhibitors. J. Enzyme Inhib. Med. Chem. 2022;37(1):1884–1902. - PMC - PubMed
-
- Khalifa M. M. Al-Karmalawy A. A. Elkaeed E. B. Nafie M. S. Tantawy M. A. Eissa I. H. Mahdy H. A. Topo II inhibition and DNA intercalation by new phthalazine-based derivatives as potent anticancer agents: design, synthesis, anti-proliferative, docking, and in vivo studies. J. Enzyme Inhib. Med. Chem. 2022;37(1):299–314. - PMC - PubMed
-
- DeVita Jr V. T. Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68(21):8643–8653. - PubMed
-
- Fidler I. J. Ellis L. M. Chemotherapeutic drugs—more really is not better. Nat. Med. 2000;6(5):500–502. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

