The Unique Genome of the Virus and Alternative Strategies for its Realization
- PMID: 37538802
- PMCID: PMC10395775
- DOI: 10.32607/actanaturae.11904
The Unique Genome of the Virus and Alternative Strategies for its Realization
Abstract
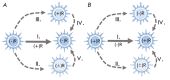
Dedicated to the 130th anniversary of Dmitry Ivanovsky's discovery of the virus kingdom as a new form of biological life. The genome of some RNA-containing viruses comprises ambipolar genes that are arranged in stacks (one above the other) encoding proteins in opposite directions. Ambipolar genes provide a new approach for developing viral diversity when virions possessing an identical genome may differ in its expression scheme (strategy) and have distinct types of progeny virions varying in the genomic RNA polarity and the composition of proteins expressed by positive- or negative-sense genes, the so-called ambipolar virions. So far, this pathway of viral genome expression remains hypothetical and hidden from us, like the dark side of the Moon, and deserves a detailed study.
Keywords: ambisense genes; genome strategy; virus classification; virus diversity.
Copyright ® 2023 National Research University Higher School of Economics.
Figures


References
-
- Ivanovsky D.I., Agriculture and Forestry. 1892;(2):108–121.
-
- Concerning the mosaic disease of the tobacco plant. 1892. Ivanowsky D., Phytopathological classics. St. Paul, MN: American Phytopathological Society. 1942;7:27–30.
-
- Zhirnov O.P., Georgiev G.P., Annals of the Russian Academy of Medical Sciences. 2017;72(1):84–86.
-
- Lvov D.K., Alkhovsky S.V., Zhirnov O.P., Probl. Virol. 2022;67(5):357–384.:10.36233/0507-4088-140. - PubMed
LinkOut - more resources
Full Text Sources
