Beyond genetics: Deciphering the impact of missense variants in CAD deficiency
- PMID: 37540500
- PMCID: PMC10838372
- DOI: 10.1002/jimd.12667
Beyond genetics: Deciphering the impact of missense variants in CAD deficiency
Abstract
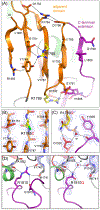
CAD is a large, 2225 amino acid multienzymatic protein required for de novo pyrimidine biosynthesis. Pathological CAD variants cause a developmental and epileptic encephalopathy which is highly responsive to uridine supplements. CAD deficiency is difficult to diagnose because symptoms are nonspecific, there is no biomarker, and the protein has over 1000 known variants. To improve diagnosis, we assessed the pathogenicity of 20 unreported missense CAD variants using a growth complementation assay that identified 11 pathogenic variants in seven affected individuals; they would benefit from uridine treatment. We also tested nine variants previously reported as pathogenic and confirmed the damaging effect of seven. However, we reclassified two variants as likely benign based on our assay, which is consistent with their long-term follow-up with uridine. We found that several computational methods are unreliable predictors of pathogenic CAD variants, so we extended the functional assay results by studying the impact of pathogenic variants at the protein level. We focused on CAD's dihydroorotase (DHO) domain because it accumulates the largest density of damaging missense changes. The atomic-resolution structures of eight DHO pathogenic variants, combined with functional and molecular dynamics analyses, provided a comprehensive structural and functional understanding of the activity, stability, and oligomerization of CAD's DHO domain. Combining our functional and protein structural analysis can help refine clinical diagnostic workflow for CAD variants in the genomics era.
Keywords: developmental and epileptic encephalopathy; dihydroorotase; epilepsy; functional validation assay; inborn metabolic disease; molecular dynamics; protein structure-function; pyrimidine metabolism; treatment; uridine; variant of uncertain significance; x-ray crystallography.
© 2023 The Authors. Journal of Inherited Metabolic Disease published by John Wiley & Sons Ltd on behalf of SSIEM.
Conflict of interest statement
Competing interest statement
Francisco del Caño-Ochoa, Bobby G. Ng, Antonio Rubio-del-Campo, Sonal Mahajan, Matthew P. Wilson, Marçal Vilar, Daisy Rymen, Paula Sánchez-Pintos, Janna Kenny, Myriam Ley Martos, Teresa Campos, Saskia B. Wortmann, Hudson H. Freeze and Santiago Ramón-Maiques declare that they have no conflict of interest related to this work. Saskia B. Wortmann receives funding from Paracelsus Medical University (PMU-FFF A-20/01/040-WOS) and ERAPERMED2019-310 – Personalized Mitochondrial Medicine (PerMiM): Optimizing diagnostics and treatment for patients with mitochondrial diseases (Austrian Science Funds (4704-B)). S.B. Wortmann declares that over the last 3 years in the area of inherited metabolic diseases she has received accommodation support from Nutricia Metabolics.
Figures



References
-
- Biesecker LG, Green RC. Diagnostic Clinical Genome and Exome Sequencing. N Engl J Med. 2014;370:2418–2425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

