FACS-based genome-wide CRISPR screens define key regulators of DNA damage signaling pathways
- PMID: 37541219
- PMCID: PMC10421629
- DOI: 10.1016/j.molcel.2023.07.004
FACS-based genome-wide CRISPR screens define key regulators of DNA damage signaling pathways
Abstract
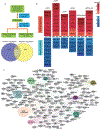
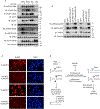
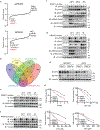
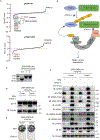
DNA damage-activated signaling pathways are critical for coordinating multiple cellular processes, which must be tightly regulated to maintain genome stability. To provide a comprehensive and unbiased perspective of DNA damage response (DDR) signaling pathways, we performed 30 fluorescence-activated cell sorting (FACS)-based genome-wide CRISPR screens in human cell lines with antibodies recognizing distinct endogenous DNA damage signaling proteins to identify critical regulators involved in DDR. We discovered that proteasome-mediated processing is an early and prerequisite event for cells to trigger camptothecin- and etoposide-induced DDR signaling. Furthermore, we identified PRMT1 and PRMT5 as modulators that regulate ATM protein level. Moreover, we discovered that GNB1L is a key regulator of DDR signaling via its role as a co-chaperone specifically regulating PIKK proteins. Collectively, these screens offer a rich resource for further investigation of DDR, which may provide insight into strategies of targeting these DDR pathways to improve therapeutic outcomes.
Keywords: DDR signaling; FACS-based CRISPR screen; GNB1L; PRMT5; antibody; proteasome.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

