Decreased TMIGD1 aggravates colitis and intestinal barrier dysfunction via the BANF1-NF-κB pathway in Crohn's disease
- PMID: 37542259
- PMCID: PMC10403950
- DOI: 10.1186/s12916-023-02989-2
Decreased TMIGD1 aggravates colitis and intestinal barrier dysfunction via the BANF1-NF-κB pathway in Crohn's disease
Abstract
Background: Disrupted intestinal epithelial barrier is one of the major causes of Crohn's disease (CD). Novel molecular targets for intestinal epithelial barrier are essential to treatment of CD. Transmembrane and immunoglobulin domain-containing protein 1 (TMIGD1) is an adhesion molecule that regulates cell adhesion, migration, and enterocyte differentiation. However, the function and mechanism of TMIGD1 in CD and intestinal epithelial barrier has rarely been studied. Furthermore, the association between TMIGD1 and the clinical features of CD remains unclear.
Methods: Transcriptome analysis on colonic mucosa from CD patients and healthy individuals were performed to identify dysregulated genes. Multi-omics integration of the 1000IBD cohort including genomics, transcriptomics of intestinal biopsies, and serum proteomics identified the association between genes and characteristics of CD. Inflammation was assessed by cytokine production in cell lines, organoids and intestinal-specific Tmigd1 knockout (Tmigd1INT-KO) mice. Epithelial barrier integrity was evaluated by trans-epithelium electrical resistance (TEER), paracellular permeability, and apical junction complex (AJC) expression. Co-immunoprecipitation, GST pull-down assays, mass spectrometry, proteomics, and transcriptome analysis were used to explore downstream mechanisms.
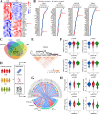
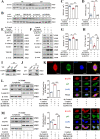
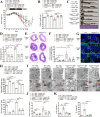
Results: Multi-omics integration suggested that TMIGD1 was negatively associated with inflammatory characteristics of CD. TMIGD1 was downregulated in inflamed intestinal mucosa of patients with CD and mice colitis models. Tmigd1INT-KO mice were more susceptible to chemically induced colitis. In epithelial cell lines and colonic organoids, TMIGD1 knockdown caused impaired intestinal barrier integrity evidenced by increased paracellular permeability and reduced TEER and AJC expression. TMIGD1 knockdown in intestinal epithelial cells also induced pro-inflammatory cytokine production. Mechanistically, TMIGD1 directly interacted with cytoplasmic BAF nuclear assembly factor 1 (BANF1) to inhibit NF-κB activation. Exogenous expression of TMIGD1 and BANF1 restored intestinal barrier function and inhibited inflammation in vitro and in vivo. TMIGD1 expression predicted response to anti-TNF treatment in patients with CD.
Conclusions: Our study demonstrated that TMIGD1 maintained intestinal barrier integrity and inactivated inflammation, and was therefore a potential therapeutic target for CD.
Keywords: BANF1; Crohn’s disease; Intestinal epithelial barrier; NF-κB pathway; TMIGD1.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declared that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

