Cardiomyocyte proliferation is suppressed by ARID1A-mediated YAP inhibition during cardiac maturation
- PMID: 37543677
- PMCID: PMC10404286
- DOI: 10.1038/s41467-023-40203-2
Cardiomyocyte proliferation is suppressed by ARID1A-mediated YAP inhibition during cardiac maturation
Abstract
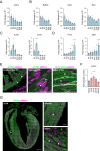
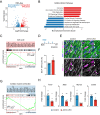
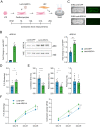
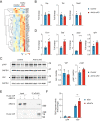
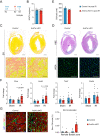
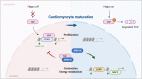
The inability of adult human cardiomyocytes to proliferate is an obstacle to efficient cardiac regeneration after injury. Understanding the mechanisms that drive postnatal cardiomyocytes to switch to a non-regenerative state is therefore of great significance. Here we show that Arid1a, a subunit of the switching defective/sucrose non-fermenting (SWI/SNF) chromatin remodeling complex, suppresses postnatal cardiomyocyte proliferation while enhancing maturation. Genome-wide transcriptome and epigenome analyses revealed that Arid1a is required for the activation of a cardiomyocyte maturation gene program by promoting DNA access to transcription factors that drive cardiomyocyte maturation. Furthermore, we show that ARID1A directly binds and inhibits the proliferation-promoting transcriptional coactivators YAP and TAZ, indicating ARID1A sequesters YAP/TAZ from their DNA-binding partner TEAD. In ischemic heart disease, Arid1a expression is enhanced in cardiomyocytes of the border zone region. Inactivation of Arid1a after ischemic injury enhanced proliferation of border zone cardiomyocytes. Our study illuminates the pivotal role of Arid1a in cardiomyocyte maturation, and uncovers Arid1a as a crucial suppressor of cardiomyocyte proliferation.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

