Identification of disulfidptosis-related subtypes, characterization of tumor microenvironment infiltration, and development of a prognosis model in colorectal cancer
- PMID: 37543978
- PMCID: PMC11796609
- DOI: 10.1007/s00432-023-05211-1
Identification of disulfidptosis-related subtypes, characterization of tumor microenvironment infiltration, and development of a prognosis model in colorectal cancer
Abstract
Background: Colorectal cancer is the second leading cause of cancer-related deaths, which imposes a significant societal burden. Regular screening and emerging molecular tumor markers have important implications for detecting the progression and development of colorectal cancer. Disulfidptosis is a newly defined type of programmed cell death triggered by abnormal accumulation of disulfide compounds in cells that stimulate disulfide stress. Currently, there is no relevant discussion on this mechanism and colorectal cancer.
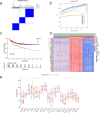
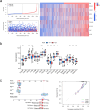
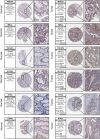
Methods: We classified the disulfidptosis-related subtypes of colorectal cancer using bioinformatics methods. Through secondary clustering of differentially expressed genes between subtypes, we identified characteristic genes of the disulfidptosis subtype, constructed a prognostic model, and searched for potential biomarkers through clinical validation.
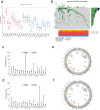
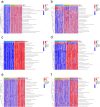
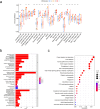
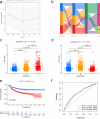
Results: Using disulfidptosis-related genes collected from the literature, we classified colorectal cancer patients from public databases into three subtypes. The differentially expressed genes between subtypes were clustered into three gene subtypes, and eight characteristic genes were screened to construct a prognostic model.
Conclusion: The disulfidptosis mechanism has important value in the classification of colorectal cancer patients, and characteristic genes selected based on this mechanism can serve as a new potential biological marker for colorectal cancer.
Keywords: Bioinformatics; Cell death; Colorectal cancer; Disulfidptosis; Prognostic model; Tumor classification.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



References
-
- Ancey PB, Contat C, Meylan E (2018) Glucose transporters in cancer—from tumor cells to the tumor microenvironment. FEBS J 285(16):2926–2943. 10.1111/febs.14577 - PubMed
-
- Augustine T, Maitra R, Zhang J, Nayak J, Goel S (2019) Sensitization of colorectal cancer to irinotecan therapy by PARP inhibitor rucaparib. Investig New Drugs 37(5):948–960. 10.1007/s10637-018-00717-9 - PubMed
-
- Bajo-Grañeras R, Crespo-Sanjuan J, García-Centeno RM, Garrote-Adrados JA, Gutierrez G, García-Tejeiro M et al (2013) Expression and potential role of apolipoprotein D on the death-survival balance of human colorectal cancer cells under oxidative stress conditions. Int J Colorectal Dis 28(6):751–766. 10.1007/s00384-012-1616-2 - PubMed
-
- Chan TC, Li CF, Ke HL, Wei YC, Shiue YL, Li CC et al (2019) High TNFAIP6 level is associated with poor prognosis of urothelial carcinomas. Urol Oncol 37(4):293.e11-293.e24. 10.1016/j.urolonc.2018.12.009 - PubMed
-
- Chen P, Li X, Zhang R, Liu S, Xiang Y, Zhang M et al (2020) Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics 10(11):5107–5119. 10.7150/thno.44705 - PMC - PubMed
LinkOut - more resources
Full Text Sources

