IFI27 Integrates Succinate and Fatty Acid Oxidation to Promote Adipocyte Thermogenic Adaption
- PMID: 37544897
- PMCID: PMC10558685
- DOI: 10.1002/advs.202301855
IFI27 Integrates Succinate and Fatty Acid Oxidation to Promote Adipocyte Thermogenic Adaption
Abstract
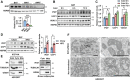
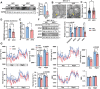
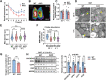
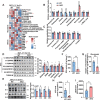
Mitochondria are the pivot organelles to control metabolism and energy homeostasis. The capacity of mitochondrial metabolic adaptions to cold stress is essential for adipocyte thermogenesis. How brown adipocytes keep mitochondrial fitness upon a challenge of cold-induced oxidative stress has not been well characterized. This manuscript shows that IFI27 plays an important role in cristae morphogenesis, keeping intact succinate dehydrogenase (SDH) function and active fatty acid oxidation to sustain thermogenesis in brown adipocytes. IFI27 protein interaction map identifies SDHB and HADHA as its binding partners. IFI27 physically links SDHB to chaperone TNF receptor associated protein 1 (TRAP1), which shields SDHB from oxidative damage-triggered degradation. Moreover, IFI27 increases hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha (HADHA) catalytic activity in β-oxidation pathway. The reduced SDH level and fatty acid oxidation in Ifi27-knockout brown fat results in impaired oxygen consumption and defective thermogenesis. Thus, IFI27 is a novel regulator of mitochondrial metabolism and thermogenesis.
Keywords: IFI27; brown adipocyte; metabolic adaption; mitochondria; thermogenesis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
