eIF3f Mediates SGOC Pathway Reprogramming by Enhancing Deubiquitinating Activity in Colorectal Cancer
- PMID: 37544925
- PMCID: PMC10520677
- DOI: 10.1002/advs.202300759
eIF3f Mediates SGOC Pathway Reprogramming by Enhancing Deubiquitinating Activity in Colorectal Cancer
Abstract
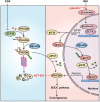
Numerous studies have demonstrated that individual proteins can moonlight. Eukaryotic Initiation translation factor 3, f subunit (eIF3f) is involved in critical biological functions; however, its role independent of protein translation in regulating colorectal cancer (CRC) is not characterized. Here, it is demonstrated that eIF3f is upregulated in CRC tumor tissues and that both Wnt and EGF signaling pathways are participating in eIF3f's oncogenic impact on targeting phosphoglycerate dehydrogenase (PHGDH) during CRC development. Mechanistically, EGF blocks FBXW7β-mediated PHGDH ubiquitination through GSK3β deactivation, and eIF3f antagonizes FBXW7β-mediated PHGDH ubiquitination through its deubiquitinating activity. Additionally, Wnt signals transcriptionally activate the expression of eIF3f, which also exerts its deubiquitinating activity toward MYC, thereby increasing MYC-mediated PHGDH transcription. Thereby, both impacts allow eIF3f to elevate the expression of PHGDH, enhancing Serine-Glycine-One-Carbon (SGOC) signaling pathway to facilitate CRC development. In summary, the study uncovers the intrinsic role and underlying molecular mechanism of eIF3f in SGOC signaling, providing novel insight into the strategies to target eIF3f-PHGDH axis in CRC.
Keywords: FBXW7; MYC; PHGDH; Wnt; colorectal cancer; eIF3f; ubiquitination.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- a) Crosby D., Bhatia S., Brindle K. M., Coussens L. M., Dive C., Emberton M., Esener S., Fitzgerald R. C., Gambhir S. S., Kuhn P., Rebbeck T. R., Balasubramanian S., Science 2022, 375, eaay9040; - PubMed
- b) Akimoto N., Ugai T., Zhong R., Hamada T., Fujiyoshi K., Giannakis M., Wu K., Cao Y., Ng K., Ogino S., Nat. Rev. Clin. Oncol. 2021, 18, 230. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2020YFA0803300/National Key R&D Program of China
- 82273133/National Natural Science Foundation of China
- 82102973/National Natural Science Foundation of China
- 81630072/National Natural Science Foundation of China
- 2023A1515030261/Natural Science Foundation of Guangdong Province
- 2021A1515012081/Natural Science Foundation of Guangdong Province
- 2020B1212060023/Open Fund of Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease
- 202206010167/Guangzhou Science and Technology Program Project
- 202201011425/Guangzhou Science and Technology Program Project
- KQTD20170810160226082/Shenzhen Municipal Government of China
- National Key Clinical Discipline
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
