This is a preprint.
Neuronal DAMPs exacerbate neurodegeneration via astrocytic RIPK3 signaling
- PMID: 37546744
- PMCID: PMC10401942
- DOI: 10.1101/2023.07.21.550097
Neuronal DAMPs exacerbate neurodegeneration via astrocytic RIPK3 signaling
Update in
-
Neuronal DAMPs exacerbate neurodegeneration via astrocytic RIPK3 signaling.JCI Insight. 2024 May 7;9(11):e177002. doi: 10.1172/jci.insight.177002. JCI Insight. 2024. PMID: 38713518 Free PMC article.
Abstract
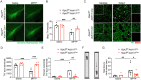
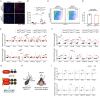
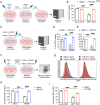
Astrocyte activation is a common feature of neurodegenerative diseases. However, the ways in which dying neurons influence the activity of astrocytes is poorly understood. RIPK3 signaling has recently been described as a key regulator of neuroinflammation, but whether this kinase mediates astrocytic responsiveness to neuronal death has not yet been studied. Here, we used the MPTP model of Parkinson's disease to show that activation of astrocytic RIPK3 drives dopaminergic cell death and axon damage. Transcriptomic profiling revealed that astrocytic RIPK3 promoted gene expression associated with neuroinflammation and movement disorders, and this coincided with significant engagement of DAMP signaling. Using human cell culture systems, we show that factors released from dying neurons signal through RAGE to induce RIPK3-dependent astrocyte activation. These findings highlight a mechanism of neuron-glia crosstalk in which neuronal death perpetuates further neurodegeneration by engaging inflammatory astrocyte activation via RIPK3.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
