This is a preprint.
Cross-site reproducibility of human cortical organoids reveals consistent cell type composition and architecture
- PMID: 37546772
- PMCID: PMC10402155
- DOI: 10.1101/2023.07.28.550873
Cross-site reproducibility of human cortical organoids reveals consistent cell type composition and architecture
Update in
-
Cross-site reproducibility of human cortical organoids reveals consistent cell type composition and architecture.Stem Cell Reports. 2024 Sep 10;19(9):1351-1367. doi: 10.1016/j.stemcr.2024.07.008. Epub 2024 Aug 22. Stem Cell Reports. 2024. PMID: 39178845 Free PMC article.
Abstract
Background: Reproducibility of human cortical organoid (hCO) phenotypes remains a concern for modeling neurodevelopmental disorders. While guided hCO protocols reproducibly generate cortical cell types in multiple cell lines at one site, variability across sites using a harmonized protocol has not yet been evaluated. We present an hCO cross-site reproducibility study examining multiple phenotypes.
Methods: Three independent research groups generated hCOs from one induced pluripotent stem cell (iPSC) line using a harmonized miniaturized spinning bioreactor protocol. scRNA-seq, 3D fluorescent imaging, phase contrast imaging, qPCR, and flow cytometry were used to characterize the 3 month differentiations across sites.
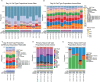
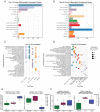
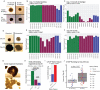
Results: In all sites, hCOs were mostly cortical progenitor and neuronal cell types in reproducible proportions with moderate to high fidelity to the in vivo brain that were consistently organized in cortical wall-like buds. Cross-site differences were detected in hCO size and morphology. Differential gene expression showed differences in metabolism and cellular stress across sites. Although iPSC culture conditions were consistent and iPSCs remained undifferentiated, primed stem cell marker expression prior to differentiation correlated with cell type proportions in hCOs.
Conclusions: We identified hCO phenotypes that are reproducible across sites using a harmonized differentiation protocol. Previously described limitations of hCO models were also reproduced including off-target differentiations, necrotic cores, and cellular stress. Improving our understanding of how stem cell states influence early hCO cell types may increase reliability of hCO differentiations. Cross-site reproducibility of hCO cell type proportions and organization lays the foundation for future collaborative prospective meta-analytic studies modeling neurodevelopmental disorders in hCOs.
Figures




References
-
- Alagappan Dhivyaa, Balan Murugabaskar, Jiang Yuhui, Cohen Rachel B., Kotenko Sergei V., and Levison Steven W.. 2013. “Egr-1 Is a Critical Regulator of EGF-Receptor-Mediated Expansion of Subventricular Zone Neural Stem Cells and Progenitors during Recovery from Hypoxia-Hypoglycemia.” ASN Neuro 5 (3): 183–93. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
