This is a preprint.
TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells
- PMID: 37546873
- PMCID: PMC10402096
- DOI: 10.1101/2023.07.25.550533
TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells
Update in
-
TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells.EMBO Rep. 2024 Jan;25(1):198-227. doi: 10.1038/s44319-023-00005-5. Epub 2023 Dec 19. EMBO Rep. 2024. PMID: 38177908 Free PMC article.
Abstract
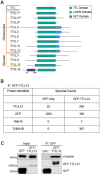
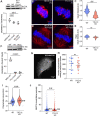
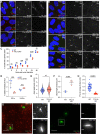
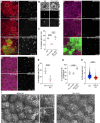
The primary cilium is a critical sensory organelle that is built of axonemal microtubules ensheathed by a ciliary membrane. In polarized epithelial cells, primary cilia reside on the apical surface and must extend these microtubules directly into the extracellular space and remain a stable structure. However, the factors regulating cross-talk between ciliation and cell polarization, as well as, axonemal microtubule growth and stabilization in polarized epithelia are not fully understood. In this study, we find TTLL12, a previously uncharacterized member of the Tubulin Tyrosine Ligase-Like (TTLL) family, localizes to the base of primary cilia and is required for cilia formation in polarized renal epithelial cells. We also show that TTLL12 directly binds to the α/β-tubulin heterodimer in vitro and regulates microtubule dynamics, stability, and post-translational modifications (PTMs). While all other TTLLs catalyze the addition of glutamate or glycine to microtubule C-terminal tails, TTLL12 uniquely affects tubulin PTMs by promoting both microtubule lysine acetylation and arginine methylation. Together, this work identifies a novel microtubule regulator and provides insight into the requirements for apical extracellular axoneme formation.
Conflict of interest statement
Conflict of Interest The authors declare that they have no conflict of interest.
Figures



References
-
- Brants J, Semenchenko K, Wasylyk C, Robert A, Carles A, Zambrano A, Pradeau-Aubreton K, Birck C, Schalken JA, Poch O et al. (2012) Tubulin tyrosine ligase like 12, a TTLL family member with SET- and TTL-like domains and roles in histone and tubulin modifications and mitosis. PLoS One 7: e51258. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
