This is a preprint.
Tracing the evolution of single-cell cancer 3D genomes: an atlas for cancer gene discovery
- PMID: 37546882
- PMCID: PMC10401964
- DOI: 10.1101/2023.07.23.550157
Tracing the evolution of single-cell cancer 3D genomes: an atlas for cancer gene discovery
Update in
-
Tracing the evolution of single-cell 3D genomes in Kras-driven cancers.Nat Genet. 2025 Aug 18. doi: 10.1038/s41588-025-02297-w. Online ahead of print. Nat Genet. 2025. PMID: 40825871
Abstract
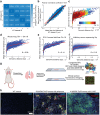
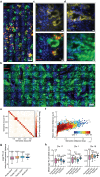
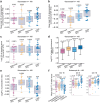
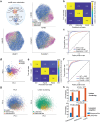
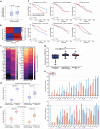
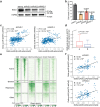
Although three-dimensional (3D) genome structures are altered in cancer cells, little is known about how these changes evolve and diversify during cancer progression. Leveraging genome-wide chromatin tracing to visualize 3D genome folding directly in tissues, we generated 3D genome cancer atlases of murine lung and pancreatic adenocarcinoma. Our data reveal stereotypical, non-monotonic, and stage-specific alterations in 3D genome folding heterogeneity, compaction, and compartmentalization as cancers progress from normal to preinvasive and ultimately to invasive tumors, discovering a potential structural bottleneck in early tumor progression. Remarkably, 3D genome architectures distinguish histologic cancer states in single cells, despite considerable cell-to-cell heterogeneity. Gene-level analyses of evolutionary changes in 3D genome compartmentalization not only showed compartment-associated genes are more homogeneously regulated, but also elucidated prognostic and dependency genes in lung adenocarcinoma and a previously unappreciated role for polycomb-group protein Rnf2 in 3D genome regulation. Our results demonstrate the utility of mapping the single-cell cancer 3D genome in tissues and illuminate its potential to identify new diagnostic, prognostic, and therapeutic biomarkers in cancer.
Conflict of interest statement
S.W., M.L., M.D.M., S.J., and S.S.A. are inventors on a patent applied for by Yale University related to this work. M.D.M. received research funding from a Genentech supported AACR grant and an honorarium from Nested Therapeutics. The remaining authors declare no competing interests.
Figures









References
-
- Zink D., Fischer A. H. & Nickerson J. A. Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 (2004). - PubMed
-
- Krijger P. H. L. & de Laat W. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 17, 771–782 (2016). - PubMed
-
- Zheng H. & Xie W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550 (2019). - PubMed
-
- Bickmore W. A. & van Steensel B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell 152, 1270–1284 (2013). - PubMed
Publication types
Grants and funding
- UH3 CA268202/CA/NCI NIH HHS/United States
- T32 GM007499/GM/NIGMS NIH HHS/United States
- P30 CA016359/CA/NCI NIH HHS/United States
- R01 LM013385/LM/NLM NIH HHS/United States
- R01 CA276108/CA/NCI NIH HHS/United States
- P50 CA196530/CA/NCI NIH HHS/United States
- U01 CA260701/CA/NCI NIH HHS/United States
- DP2 CA248136/CA/NCI NIH HHS/United States
- R01 CA292936/CA/NCI NIH HHS/United States
- DP2 GM137414/GM/NIGMS NIH HHS/United States
- T32 CA193200/CA/NCI NIH HHS/United States
- R01 HG012969/HG/NHGRI NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
- R33 CA251037/CA/NCI NIH HHS/United States
- R01 HG011245/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
