This is a preprint.
Pathogenic CANVAS (AAGGG)n repeats stall DNA replication due to the formation of alternative DNA structures
- PMID: 37546920
- PMCID: PMC10402041
- DOI: 10.1101/2023.07.25.550509
Pathogenic CANVAS (AAGGG)n repeats stall DNA replication due to the formation of alternative DNA structures
Update in
-
Pathogenic CANVAS (AAGGG)n repeats stall DNA replication due to the formation of alternative DNA structures.Nucleic Acids Res. 2024 May 8;52(8):4361-4374. doi: 10.1093/nar/gkae124. Nucleic Acids Res. 2024. PMID: 38381906 Free PMC article.
Abstract
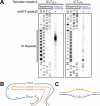
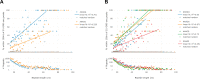
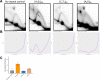
CANVAS is a recently characterized repeat expansion disease, most commonly caused by homozygous expansions of an intronic (A2G3)n repeat in the RFC1 gene. There are a multitude of repeat motifs found in the human population at this locus, some of which are pathogenic and others benign. In this study, we conducted structure-functional analyses of the main pathogenic (A2G3)n and the main nonpathogenic (A4G)n repeats. We found that the pathogenic, but not the nonpathogenic, repeat presents a potent, orientation-dependent impediment to DNA polymerization in vitro. The pattern of the polymerization blockage is consistent with triplex or quadruplex formation in the presence of magnesium or potassium ions, respectively. Chemical probing of both repeats in supercoiled DNA reveals triplex H-DNA formation by the pathogenic repeat. Consistently, bioinformatic analysis of the S1-END-seq data from human cell lines shows preferential H-DNA formation genome-wide by (A2G3)n motifs over (A4G)n motifs in vivo. Finally, the pathogenic, but not the non-pathogenic, repeat stalls replication fork progression in yeast and human cells. We hypothesize that CANVAS-causing (A2G3)n repeat represents a challenge to genome stability by folding into alternative DNA structures that stall DNA replication.
Figures


References
-
- Arteche-López A. et al. New Cerebellar Ataxia, Neuropathy, Vestibular Areflexia Syndrome cases are caused by the presence of a nonsense variant in compound heterozygosity with the pathogenic repeat expansion in the RFC1 gene. Clin Genet 103, 236–241 (2023). - PubMed
-
- Cortese A. et al. Cerebellar ataxia, neuropathy and vestibular areflexia syndrome (CANVAS): genetic and clinical aspects. Pract Neurol 22, 14–18 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
