This is a preprint.
APOE4/4 is linked to damaging lipid droplets in Alzheimer's microglia
- PMID: 37546938
- PMCID: PMC10401952
- DOI: 10.1101/2023.07.21.549930
APOE4/4 is linked to damaging lipid droplets in Alzheimer's microglia
Update in
-
APOE4/4 is linked to damaging lipid droplets in Alzheimer's disease microglia.Nature. 2024 Apr;628(8006):154-161. doi: 10.1038/s41586-024-07185-7. Epub 2024 Mar 13. Nature. 2024. PMID: 38480892 Free PMC article.
Abstract
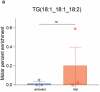
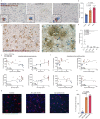
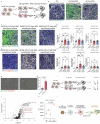
Several genetic risk factors for Alzheimer's Disease (AD) implicate genes involved in lipid metabolism and many of these lipid genes are highly expressed in glial cells. However, the relationship between lipid metabolism in glia and AD pathology remains poorly understood. Through single-nucleus RNA-sequencing of AD brain tissue, we have identified a microglial state defined by the expression of the lipid droplet (LD) associated enzyme ACSL1 with ACSL1-positive microglia most abundant in AD patients with the APOE4/4 genotype. In human iPSC-derived microglia (iMG) fibrillar Aβ (fAβ) induces ACSL1 expression, triglyceride synthesis, and LD accumulation in an APOE-dependent manner. Additionally, conditioned media from LD-containing microglia leads to Tau phosphorylation and neurotoxicity in an APOE-dependent manner. Our findings suggest a link between genetic risk factors for AD with microglial LD accumulation and neurotoxic microglial-derived factors, potentially providing novel therapeutic strategies for AD.
Conflict of interest statement
Competing financial interests T.G.B. is a paid consultant to Aprinoia Therapeutics and Biogen. E.M.R. is a scientific advisor to Alzheon, Aural Analytics, Denali, Retromer Therapeutics, and Vaxxinity and a co-founder and advisor to ALZPath. The other authors declare no competing financial interests.
Figures









References
-
- Alzheimer A. Über eine eigenartige Erkrankung der Hirnrinde. Allg. Zeitschrift fur Psychiatr. und Psych. Medizin 146–148 (1907).
Methods references
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
