This is a preprint.
An allelic series at the EDNRB2 locus controls diverse piebalding patterns in the domestic pigeon
- PMID: 37546953
- PMCID: PMC10402103
- DOI: 10.1101/2023.07.26.550625
An allelic series at the EDNRB2 locus controls diverse piebalding patterns in the domestic pigeon
Update in
-
An allelic series at the EDNRB2 locus controls diverse piebalding patterns in the domestic pigeon.PLoS Genet. 2023 Oct 20;19(10):e1010880. doi: 10.1371/journal.pgen.1010880. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37862332 Free PMC article.
Abstract
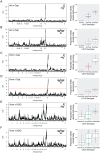
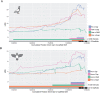
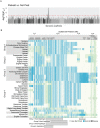
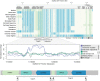
Variation in pigment patterns within and among vertebrate species reflects underlying changes in cell migration and function that can impact health, reproductive success, and survival. The domestic pigeon (Columba livia) is an exceptional model for understanding the genetic changes that give rise to diverse pigment patterns, as selective breeding has given rise to hundreds of breeds with extensive variation in plumage color and pattern. Here, we map the genetic architecture of a suite of pigmentation phenotypes known as piebalding. Piebalding is characterized by patches of pigmented and non-pigmented feathers, and these plumage patterns are often breed-specific and stable across generations. Using a combination of quantitative trait locus mapping in F2 laboratory crosses and genome-wide association analysis, we identify a locus associated with piebalding across many pigeon breeds. This shared locus harbors a candidate gene, EDNRB2, that is a known regulator of pigment cell migration, proliferation, and survival. We discover multiple distinct haplotypes at the EDNRB2 locus in piebald pigeons, which include a mix of protein-coding, noncoding, and structural variants that are associated with depigmentation in specific plumage regions. These results identify a role for EDNRB2 in pigment patterning in the domestic pigeon, and highlight how repeated selection at a single locus can generate a diverse array of stable and heritable pigment patterns.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
