This is a preprint.
The lncRNA Malat1 Inhibits miR-15/16 to Enhance Cytotoxic T Cell Activation and Memory Cell Formation
- PMID: 37547023
- PMCID: PMC10401941
- DOI: 10.1101/2023.04.14.536843
The lncRNA Malat1 Inhibits miR-15/16 to Enhance Cytotoxic T Cell Activation and Memory Cell Formation
Update in
-
The lncRNA Malat1 inhibits miR-15/16 to enhance cytotoxic T cell activation and memory cell formation.Elife. 2023 Dec 21;12:RP87900. doi: 10.7554/eLife.87900. Elife. 2023. PMID: 38127070 Free PMC article.
Abstract
Proper activation of cytotoxic T cells via the T cell receptor and the costimulatory receptor CD28 is essential for adaptive immunity against viruses, many intracellular bacteria and cancers. Through biochemical analysis of RNA:protein interactions, we uncovered a non-coding RNA circuit regulating activation and differentiation of cytotoxic T cells composed of the long non-coding RNA Malat1 (Metastasis Associated Lung Adenocarcinoma Transcript 1) and the microRNA family miR-15/16. miR-15/16 is a widely and highly expressed tumor suppressor miRNA family important for cell proliferation and survival. miR-15/16 also play important roles in T cell responses to viral infection, including the regulation of antigen-specific T cell expansion and T cell memory. Comparative Argonaute-2 high throughput sequencing of crosslinking immunoprecipitation (Ago2 HITS-CLIP, or AHC) combined with gene expression profiling in normal and miR-15/16-deficient T cells revealed a large network of several hundred direct miR-15/16 target mRNAs, many with functional relevance for T cell activation, survival and memory formation. Among these targets, the long non-coding RNA Malat1 contained the largest absolute magnitude miR-15/16-dependent AHC peak in T cells. This binding site was also among the strongest lncRNA:miRNA interactions detected in the T cell transcriptome. We used CRISPR targeting with homology directed repair to generate mice with a 5-nucleotide mutation in the miR-15/16 binding site in Malat1. This mutation interrupted Malat1:miR-15/16 interaction, and enhanced the repression of other miR-15/16 target genes, including CD28. Interrupting Malat1 interaction with miR-15/16 decreased cytotoxic T cell activation, including the expression of IL-2 and a broader CD28-responsive gene program. Accordingly, Malat1 mutation diminished memory cell persistence following LCMV Armstrong and Listeria monocytogenes infection. This study marks a significant advance in the study of long noncoding RNAs in the immune system by ascribing cell-intrinsic, sequence-specific in vivo function to Malat1. These findings have implications for T cell-mediated autoimmune diseases, antiviral and anti-tumor immunity, as well as lung adenocarcinoma and other malignancies where Malat1 is overexpressed.
Conflict of interest statement
A.M. is a cofounder of Arsenal Biosciences, Spotlight Therapeutics, and Survey Genomics, serves on the boards of directors at Spotlight Therapeutics and Survey Genomics, is a board observer (and former member of the board of directors) at Arsenal Biosciences, is a member of the scientific advisory boards of Arsenal Biosciences, Spotlight Therapeutics, Survey Genomics, NewLimit, Amgen and Tenaya, owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, PACT Pharma, and Tenaya and has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, 23andMe, PACT Pharma, Juno Therapeutics, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. The Marson laboratory has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead and Anthem. M.H.S. is founder and a board member of Teiko.bio and has received a speaking honorarium from Fluidigm Inc., has served as a consultant for Five Prime, Ono, January, Earli, Astellas, and Indaptus, and has received research funding from Roche/Genentech, Bristol Myers Squibb, Valitor, and Pfizer. J.D.G. is now an employee of Arsenal Biosciences.
Figures

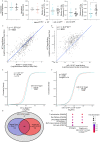





References
-
- Bachmann MF, Barner M, Viola A, Kopf M. 1999. Distinct kinetics of cytokine production and cytolysis in effector and memory T cells after viral infection. Eur J Immunol 29:291–299. - PubMed
-
- Badovinac VP, Porter BB, Harty JT. 2002. Programmed contraction of CD8(+) T cells after infection. Nat Immunol 3:619–626. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
