Genome analyses of species A rotavirus isolated from various mammalian hosts in Northern Ireland during 2013-2016
- PMID: 37547380
- PMCID: PMC10403756
- DOI: 10.1093/ve/vead039
Genome analyses of species A rotavirus isolated from various mammalian hosts in Northern Ireland during 2013-2016
Abstract
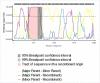
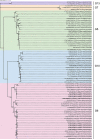
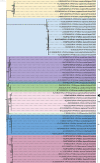
Rotavirus group A (RVA) is the most important cause of acute diarrhoea and severe dehydration in young mammals. Infection in livestock is associated with significant mortality and economic losses and, together with wildlife reservoirs, acts as a potential source of zoonotic transmission. Therefore, molecular surveillance of circulating RVA strains in animal species is necessary to assess the risks posed to humans and their livestock. An RVA molecular epidemiological surveillance study on clinically diseased livestock species revealed high prevalence in cattle and pigs (31 per cent and 18 per cent, respectively) with significant phylogenetic diversity including a novel and divergent ovine artiodactyl DS-1-like constellation G10-P[15]-I2-R2-C2-M2-A11-N2-T6-E2-H3. An RVA gene reassortment occurred in an RVA asymptomatic pig and identified as a G5-P[13] strain, and a non-structural protein (NSP)2 gene had intergenomically reassorted with a human RVA strain (reverse zoonosis) and possessed a novel NSP4 enterotoxin E9 which may relate to the asymptomatic RVA infection. Analysis of a novel sheep G10-P[15] strain viral protein 4 gene imparts a putative homologous intergenic and interspecies recombination event, subsequently creating the new P[15] divergent lineage. While surveillance across a wider range of wildlife and exotic species identified generally negative or low prevalence, a novel RVA interspecies transmission in a non-indigenous pudu deer (zoo origin) with the constellation of G6-P[11]12-R2-C2-M2-A3-N2-T6-E2-H3 was detected at a viral load of 11.1 log10 copies/gram. The detection of novel emerging strains, interspecies reassortment, interspecies infection, and recombination of RVA circulating in animal livestock and wildlife reservoirs is of paramount importance to the RVA epidemiology and evolution for the One Health approach and post-human vaccine introduction era where highly virulent animal RVA genotypes have the potential to be zoonotically transmitted.
Keywords: NGS; WGS; interspecies; phylogenetics; prevalence; reassortment; recombination; rotavirus group A; surveillance.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Multispecies reassortant bovine rotavirus strain carries a novel simian G3-like VP7 genotype.Infect Genet Evol. 2016 Jul;41:63-72. doi: 10.1016/j.meegid.2016.03.023. Epub 2016 Mar 28. Infect Genet Evol. 2016. PMID: 27033751
-
Genomic revelations: investigating rotavirus a presence in wild ruminants and its zoonotic potential.Front Vet Sci. 2024 Aug 15;11:1429654. doi: 10.3389/fvets.2024.1429654. eCollection 2024. Front Vet Sci. 2024. PMID: 39211480 Free PMC article.
-
Whole genome sequence analyses of three African bovine rotaviruses reveal that they emerged through multiple reassortment events between rotaviruses from different mammalian species.Vet Microbiol. 2012 Sep 14;159(1-2):245-50. doi: 10.1016/j.vetmic.2012.03.040. Epub 2012 Apr 6. Vet Microbiol. 2012. PMID: 22541163
-
Zoonotic transmission of rotavirus: surveillance and control.Expert Rev Anti Infect Ther. 2015;13(11):1337-50. doi: 10.1586/14787210.2015.1089171. Epub 2015 Oct 1. Expert Rev Anti Infect Ther. 2015. PMID: 26428261 Review.
-
Zoonotic RVA: State of the Art and Distribution in the Animal World.Viruses. 2022 Nov 18;14(11):2554. doi: 10.3390/v14112554. Viruses. 2022. PMID: 36423163 Free PMC article. Review.
Cited by
-
Full genome characterization of a Kenyan G8P[14] rotavirus strain suggests artiodactyl-to-human zoonotic transmission.Trop Med Health. 2025 Jun 16;53(1):82. doi: 10.1186/s41182-025-00759-9. Trop Med Health. 2025. PMID: 40524227 Free PMC article.
References
-
- Adah M. I. et al. (2002) ‘Detection of Human Rotavirus in Faeces from Diarrhoeic Calves in North-east Nigeria’, Tropical Animal Health and Production, 34: 1–6. - PubMed
-
- Adams W. R., and Kraft L. M. (1963) ‘Epizootic Diarrhea of Infant Mice: Identification of the Etiologic Agent’, Science, 141: 359–60. - PubMed
-
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

