Missingness adapted group informed clustered (MAGIC)-LASSO: a novel paradigm for phenotype prediction to improve power for genetic loci discovery
- PMID: 37547462
- PMCID: PMC10399453
- DOI: 10.3389/fgene.2023.1162690
Missingness adapted group informed clustered (MAGIC)-LASSO: a novel paradigm for phenotype prediction to improve power for genetic loci discovery
Abstract
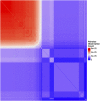
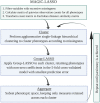
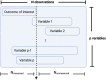
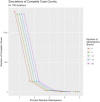
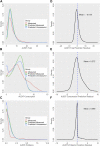
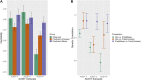
Introduction: The availability of large-scale biobanks linking genetic data, rich phenotypes, and biological measures is a powerful opportunity for scientific discovery. However, real-world collections frequently have extensive missingness. While missing data prediction is possible, performance is significantly impaired by block-wise missingness inherent to many biobanks. Methods: To address this, we developed Missingness Adapted Group-wise Informed Clustered (MAGIC)-LASSO which performs hierarchical clustering of variables based on missingness followed by sequential Group LASSO within clusters. Variables are pre-filtered for missingness and balance between training and target sets with final models built using stepwise inclusion of features ranked by completeness. This research has been conducted using the UK Biobank (n > 500 k) to predict unmeasured Alcohol Use Disorders Identification Test (AUDIT) scores. Results: The phenotypic correlation between measured and predicted total score was 0.67 while genetic correlations between independent subjects was high >0.86. Discussion: Phenotypic and genetic correlations in real data application, as well as simulations, demonstrate the method has significant accuracy and utility for increasing power for genetic loci discovery.
Keywords: GWAS; LASSO; UK biobank; alcohol consumption; genetics; machine learning; missingness; prediction.
Copyright © 2023 Gentry, Kirkpatrick, Peterson and Webb.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bates D., Maechler M. (2019). Matrix: Sparse and dense matrix classes and methods. Available at: https://CRAN.R-project.org/package=Matrix .
-
- Breheny P., Zeng Y. (2022). Grpreg: Regularization paths for regression models with grouped covariates. Available at: https://cran.r-project.org/web/packages/grpreg/index.html .
-
- Bulik-Sullivan B. K., Loh P-R., Finucane H. K., Ripke S., Yang J. Schizophrenia Working Group of the Psychiatric Genomics Consortium (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47 (3), 291–295. Nature Publishing Group. 10.1038/ng.3211 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

