This is a preprint.
Weakly Supervised AI for Efficient Analysis of 3D Pathology Samples
- PMID: 37547660
- PMCID: PMC10402184
Weakly Supervised AI for Efficient Analysis of 3D Pathology Samples
Update in
-
Analysis of 3D pathology samples using weakly supervised AI.Cell. 2024 May 9;187(10):2502-2520.e17. doi: 10.1016/j.cell.2024.03.035. Cell. 2024. PMID: 38729110 Free PMC article.
Abstract
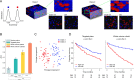
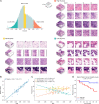
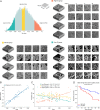
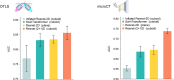
Human tissue consists of complex structures that display a diversity of morphologies, forming a tissue microenvironment that is, by nature, three-dimensional (3D). However, the current standard-of-care involves slicing 3D tissue specimens into two-dimensional (2D) sections and selecting a few for microscopic evaluation1,2, with concomitant risks of sampling bias and misdiagnosis3-6. To this end, there have been intense efforts to capture 3D tissue morphology and transition to 3D pathology, with the development of multiple high-resolution 3D imaging modalities7-18. However, these tools have had little translation to clinical practice as manual evaluation of such large data by pathologists is impractical and there is a lack of computational platforms that can efficiently process the 3D images and provide patient-level clinical insights. Here we present Modality-Agnostic Multiple instance learning for volumetric Block Analysis (MAMBA), a deep-learning-based platform for processing 3D tissue images from diverse imaging modalities and predicting patient outcomes. Archived prostate cancer specimens were imaged with open-top light-sheet microscopy12-14 or microcomputed tomography15,16 and the resulting 3D datasets were used to train risk-stratification networks based on 5-year biochemical recurrence outcomes via MAMBA. With the 3D block-based approach, MAMBA achieves an area under the receiver operating characteristic curve (AUC) of 0.86 and 0.74, superior to 2D traditional single-slice-based prognostication (AUC of 0.79 and 0.57), suggesting superior prognostication with 3D morphological features. Further analyses reveal that the incorporation of greater tissue volume improves prognostic performance and mitigates risk prediction variability from sampling bias, suggesting that there is value in capturing larger extents of spatially heterogeneous 3D morphology. With the rapid growth and adoption of 3D spatial biology and pathology techniques by researchers and clinicians, MAMBA provides a general and efficient framework for 3D weakly supervised learning for clinical decision support and can help to reveal novel 3D morphological biomarkers for prognosis and therapeutic response.
Conflict of interest statement
Competing Interests J.T.C.L. is a co-founder and board member of Alpenglow Biosciences, Inc., which has licensed the OTLS microscopy portfolio developed in his lab at the University of Washington.
Figures









References
-
- Farahani N., Parwani A. V., Pantanowitz L. et al. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathol Lab Med Int 7, 4321 (2015).
-
- King C. R. & Long J. P. Prostate biopsy grading errors: a sampling problem? International journal of cancer 90, 326–330 (2000). - PubMed
-
- Mehra K. K. et al. The impact of tissue block sampling on the detection of p53 signatures in fallopian tubes from women with BRCA 1 or 2 mutations (BRCA+) and controls. Modern Pathology 24, 152–156 (2011). - PubMed
-
- Olson S. M., Hussaini M. & Lewis J. S. Frozen section analysis of margins for head and neck tumor resections: reduction of sampling errors with a third histologic level. Modern Pathology 24, 665–670 (2011). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
