Identification of microbial metabolites that accelerate the ubiquitin-dependent degradation of c-Myc
- PMID: 37547761
- PMCID: PMC10398403
- DOI: 10.32604/or.2023.030248
Identification of microbial metabolites that accelerate the ubiquitin-dependent degradation of c-Myc
Abstract
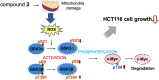
Myc belongs to a family of proto-oncogenes that encode transcription factors. The overexpression of c-Myc causes many types of cancers. Recently, we established a system for screening c-Myc inhibitors and identified antimycin A by screening the RIKEN NPDepo chemical library. The specific mechanism of promoting tumor cell metastasis by high c-Myc expression remains to be explained. In this study, we screened approximately 5,600 microbial extracts using this system and identified a broth prepared from Streptomyces sp. RK19-A0402 strongly inhibits c-Myc transcriptional activity. After purification of the hit broth, we identified compounds closely related to the aglycone of cytovaricin and had a structure similar to that of oligomycin A. Similar to oligomycin A, the hit compounds inhibited mitochondrial complex V. The mitochondria dysfunction caused by the compounds induced the production of reactive oxygen species (ROS), and the ROS activated GSK3α/β that phosphorylated c-Myc for ubiquitination. This study provides a successful screening strategy for identifying natural products as potential c-Myc inhibitors as potential anticancer agents.
Keywords: High-throughput screening; Phosphorylation; Reactive oxygen species.
© 2023 Liu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest to report regarding the present study.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
