Effect of image resolution on automated classification of chest X-rays
- PMID: 37547812
- PMCID: PMC10403240
- DOI: 10.1117/1.JMI.10.4.044503
Effect of image resolution on automated classification of chest X-rays
Abstract
Purpose: Deep learning (DL) models have received much attention lately for their ability to achieve expert-level performance on the accurate automated analysis of chest X-rays (CXRs). Recently available public CXR datasets include high resolution images, but state-of-the-art models are trained on reduced size images due to limitations on graphics processing unit memory and training time. As computing hardware continues to advance, it has become feasible to train deep convolutional neural networks on high-resolution images without sacrificing detail by downscaling. This study examines the effect of increased resolution on CXR classification performance.
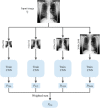
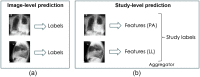
Approach: We used the publicly available MIMIC-CXR-JPG dataset, comprising 377,110 high resolution CXR images for this study. We applied image downscaling from native resolution to , , , and and then we used the DenseNet121 and EfficientNet-B4 DL models to evaluate clinical task performance using these four downscaled image resolutions.
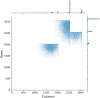
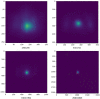
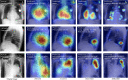
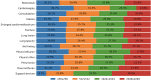
Results: We find that while some clinical findings are more reliably labeled using high resolutions, many other findings are actually labeled better using downscaled inputs. We qualitatively verify that tasks requiring a large receptive field are better suited to downscaled low resolution input images, by inspecting effective receptive fields and class activation maps of trained models. Finally, we show that stacking an ensemble across resolutions outperforms each individual learner at all input resolutions while providing interpretable scale weights, indicating that diverse information is extracted across resolutions.
Conclusions: This study suggests that instead of focusing solely on the finest image resolutions, multi-scale features should be emphasized for information extraction from high-resolution CXRs.
Keywords: chest X-ray; deep learning; image resolution; multitask classification; receptive field.
© 2023 Society of Photo-Optical Instrumentation Engineers (SPIE).
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

