A Transcriptomic Classifier Model Identifies High-Risk Endotypes in a Prospective Study of Sepsis in Uganda
- PMID: 37548511
- PMCID: PMC10847381
- DOI: 10.1097/CCM.0000000000006023
A Transcriptomic Classifier Model Identifies High-Risk Endotypes in a Prospective Study of Sepsis in Uganda
Abstract
Objectives: In high-income countries (HICs), sepsis endotypes defined by distinct pathobiological mechanisms, mortality risks, and responses to corticosteroid treatment have been identified using blood transcriptomics. The generalizability of these endotypes to low-income and middle-income countries (LMICs), where the global sepsis burden is concentrated, is unknown. We sought to determine the prevalence, prognostic relevance, and immunopathological features of HIC-derived transcriptomic sepsis endotypes in sub-Saharan Africa.
Design: Prospective cohort study.
Setting: Public referral hospital in Uganda.
Patients: Adults ( n = 128) hospitalized with suspected sepsis.
Interventions: None.
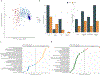
Measurements and main results: Using whole-blood RNA sequencing data, we applied 19-gene and 7-gene classifiers derived and validated in HICs (SepstratifieR) to assign patients to one of three sepsis response signatures (SRS). The 19-gene classifier assigned 30 (23.4%), 92 (71.9%), and 6 (4.7%) patients to SRS-1, SRS-2, and SRS-3, respectively, the latter of which is designed to capture individuals transcriptionally closest to health. SRS-1 was defined biologically by proinflammatory innate immune activation and suppressed natural killer-cell, T-cell, and B-cell immunity, whereas SRS-2 was characterized by dampened innate immune activation, preserved lymphocyte immunity, and suppressed transcriptional responses to corticosteroids. Patients assigned to SRS-1 were predominantly (80.0% [24/30]) persons living with HIV with advanced immunosuppression and frequent tuberculosis. Mortality at 30-days differed significantly by endotype and was highest (48.1%) in SRS-1. Agreement between 19-gene and 7-gene SRS assignments was poor (Cohen's kappa 0.11). Patient stratification was suboptimal using the 7-gene classifier with 15.1% (8/53) of individuals assigned to SRS-3 deceased at 30-days.
Conclusions: Sepsis endotypes derived in HICs share biological and clinical features with those identified in sub-Saharan Africa, with major differences in host-pathogen profiles. Our findings highlight the importance of context-specific sepsis endotyping, the generalizability of conserved biological signatures of critical illness across disparate settings, and opportunities to develop more pathobiologically informed sepsis treatment strategies in LMICs.
Copyright © 2023 by the Society of Critical Care Medicine and Wolters Kluwer Health, Inc. All Rights Reserved.
Conflict of interest statement
This work was supported by the National Center for Advancing Translational Sciences (UL1TR001873 to Columbia University, subaward to Dr. O’Donnell), the National Institute of Allergy and Infectious Diseases (K23AI163364 to Dr. Cummings), and the MakCHS-Berkeley-Yale Pulmonary Complications of AIDS Research Training Program (D43TW009607, subaward to Dr. Bakamutumaho) from the Fogarty International Center, National Institutes of Health (NIH). Additional support was provided by the Stony Wold-Herbert Fund (Dr. Cummings), Potts Memorial Foundation (Dr. Cummings), Thrasher Research Fund (Dr. Cummings), Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene (Dr. Cummings), and DELTAS Africa Initiative (subaward to Drs. Cummings and Bakamutumaho; grant no. 107743). Drs. Cummings, Bakamutumaho, Owor, Namulondo, Nayiga, Kyebambe, Lutwama, and Lipkin received support for article research from the NIH. Dr. Lipkin disclosed government work. The remaining authors have disclosed that they do not have any potential conflicts of interest.
Figures
References
-
- Maslove DM, Tang B, Shankar-Hari M, et al. Redefining critical illness. Nat Med. 2022;28:1141–1148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

