Cell Free Bacteriophage Synthesis from Engineered Strains Improves Yield
- PMID: 37548960
- PMCID: PMC10443043
- DOI: 10.1021/acssynbio.3c00239
Cell Free Bacteriophage Synthesis from Engineered Strains Improves Yield
Abstract
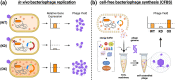
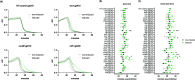
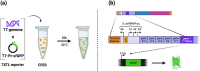
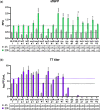
Phage therapy to treat life-threatening drug-resistant infections has been hampered by technical challenges in phage production. Cell-free bacteriophage synthesis (CFBS) can overcome the limitations of standard phage production methods by manufacturing phage virions in vitro. CFBS mimics intracellular phage assembly using transcription/translation machinery (TXTL) harvested from bacterial lysates and combined with reagents to synthesize proteins encoded by a phage genomic DNA template. These systems may enable rapid phage production and engineering to accelerate phages from bench-to-bedside. TXTL harvested from wild type or commonly used bacterial strains was not optimized for bacteriophage production. Here, we demonstrate that TXTL from genetically modified E. coli BL21 can be used to enhance phage T7 yields in vitro by CFBS. Expression of 18 E. coli BL21 genes was manipulated by inducible CRISPR interference (CRISPRi) mediated by nuclease deficient Cas12a from F. novicida (dFnCas12a) to identify genes implicated in T7 propagation as positive or negative effectors. Genes shown to have a significant effect were overexpressed (positive effectors) or repressed (negative effectors) to modify the genetic background of TXTL harvested for CFBS. Phage T7 CFBS yields were improved by up to 10-fold in vitro through overexpression of translation initiation factor IF-3 (infC) and small RNAs OxyS and CyaR and by repression of RecC subunit exonuclease RecBCD. Continued improvement of CFBS will mitigate phage manufacturing bottlenecks and lower hurdles to widespread adoption of phage therapy.
Keywords: CRISRPi; T7; TX-TL; cell-free bacteriophage synthesis (CFBS); cell-free expression systems; gene expression.
Conflict of interest statement
The authors declare the following competing financial interest(s): A provisional patent has been filed by RB protecting the enhanced TXTL for cell-free bacteriophage synthesis. Prv Appln. No. 63/332,901.
Figures


Similar articles
-
PHEIGES: all-cell-free phage synthesis and selection from engineered genomes.Nat Commun. 2024 Mar 12;15(1):2223. doi: 10.1038/s41467-024-46585-1. Nat Commun. 2024. PMID: 38472230 Free PMC article.
-
Construction of Leaderless-Bacteriocin-Producing Bacteriophage Targeting E. coli and Neighboring Gram-Positive Pathogens.Microbiol Spectr. 2021 Sep 3;9(1):e0014121. doi: 10.1128/Spectrum.00141-21. Epub 2021 Jul 14. Microbiol Spectr. 2021. PMID: 34259542 Free PMC article.
-
Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning.Elife. 2021 Mar 9;10:e63775. doi: 10.7554/eLife.63775. Elife. 2021. PMID: 33687327 Free PMC article.
-
T7 Phage as an Emerging Nanobiomaterial with Genetically Tunable Target Specificity.Adv Sci (Weinh). 2022 Feb;9(4):e2103645. doi: 10.1002/advs.202103645. Epub 2021 Dec 16. Adv Sci (Weinh). 2022. PMID: 34914854 Free PMC article. Review.
-
Recent trends in T7 phage application in diagnosis and treatment of various diseases.Int Immunopharmacol. 2022 Sep;110:109071. doi: 10.1016/j.intimp.2022.109071. Epub 2022 Jul 26. Int Immunopharmacol. 2022. PMID: 35978521 Review.
Cited by
-
Characterization and purification of Pseudomonas aeruginosa phages for the treatment of canine infections.BMC Microbiol. 2025 May 14;25(1):289. doi: 10.1186/s12866-025-04005-4. BMC Microbiol. 2025. PMID: 40369432 Free PMC article.
-
Synthetic Biology-Based Engineering Living Therapeutics for Antimicrobial Application.Exploration (Beijing). 2025 Apr 3;5(4):e20240045. doi: 10.1002/EXP.20240045. eCollection 2025 Aug. Exploration (Beijing). 2025. PMID: 40873635 Free PMC article.
-
The Biotechnological Application of Bacteriophages: What to Do and Where to Go in the Middle of the Post-Antibiotic Era.Microorganisms. 2023 Sep 13;11(9):2311. doi: 10.3390/microorganisms11092311. Microorganisms. 2023. PMID: 37764155 Free PMC article. Review.
-
Profiling expression strategies for a type III polyketide synthase in a lysate-based, cell-free system.Sci Rep. 2024 Jun 6;14(1):12983. doi: 10.1038/s41598-024-61376-w. Sci Rep. 2024. PMID: 38839808 Free PMC article.
-
Between Centralization and Fragmentation: The Past, Present, and Future of Phage Collections.Phage (New Rochelle). 2024 Mar 18;5(1):22-29. doi: 10.1089/phage.2023.0043. eCollection 2024 Mar. Phage (New Rochelle). 2024. PMID: 40114810 Free PMC article. Review.
References
-
- Antibiotic Resistance Threats in the United States; US Centers for Disease Control: 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

