Effective mechanical potential of cell-cell interaction explains three-dimensional morphologies during early embryogenesis
- PMID: 37549166
- PMCID: PMC10434874
- DOI: 10.1371/journal.pcbi.1011306
Effective mechanical potential of cell-cell interaction explains three-dimensional morphologies during early embryogenesis
Abstract
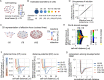
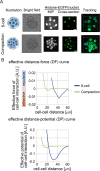
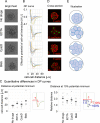
Mechanical forces are critical for the emergence of diverse three-dimensional morphologies of multicellular systems. However, it remains unclear what kind of mechanical parameters at cellular level substantially contribute to tissue morphologies. This is largely due to technical limitations of live measurements of cellular forces. Here we developed a framework for inferring and modeling mechanical forces of cell-cell interactions. First, by analogy to coarse-grained models in molecular and colloidal sciences, we approximated cells as particles, where mean forces (i.e. effective forces) of pairwise cell-cell interactions are considered. Then, the forces were statistically inferred by fitting the mathematical model to cell tracking data. This method was validated by using synthetic cell tracking data resembling various in vivo situations. Application of our method to the cells in the early embryos of mice and the nematode Caenorhabditis elegans revealed that cell-cell interaction forces can be written as a pairwise potential energy in a manner dependent on cell-cell distances. Importantly, the profiles of the pairwise potentials were quantitatively different among species and embryonic stages, and the quantitative differences correctly described the differences of their morphological features such as spherical vs. distorted cell aggregates, and tightly vs. non-tightly assembled aggregates. We conclude that the effective pairwise potential of cell-cell interactions is a live measurable parameter whose quantitative differences can be a parameter describing three-dimensional tissue morphologies.
Copyright: © 2023 Koyama et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

