Targeting STING oligomerization with small-molecule inhibitors
- PMID: 37549268
- PMCID: PMC10434303
- DOI: 10.1073/pnas.2305420120
Targeting STING oligomerization with small-molecule inhibitors
Abstract
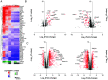
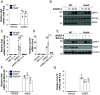
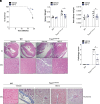
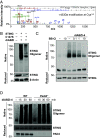
Stimulator of interferon genes (STING) is an essential adaptor protein required for the inflammatory response to cytosolic DNA. dsDNA activates cGAS to generate cGAMP, which binds and activates STING triggering a conformational change, oligomerization, and the IRF3- and NFκB-dependent transcription of type I Interferons (IFNs) and inflammatory cytokines, as well as the activation of autophagy. Aberrant activation of STING is now linked to a growing number of both rare as well as common chronic inflammatory diseases. Here, we identify and characterize a potent small-molecule inhibitor of STING. This compound, BB-Cl-amidine inhibits STING signaling and production of type I IFNs, IFN-stimulated genes (ISGs) and NFκB-dependent cytokines, but not other pattern recognition receptors. In vivo, BB-Cl-amidine alleviated pathology resulting from accrual of cytosolic DNA in Trex-1 mutant mice. Mechanistically BB-Cl-amidine inhibited STING oligomerization through modification of Cys148. Collectively, our work uncovers an approach to inhibit STING activation and highlights the potential of this strategy for the treatment of STING-driven inflammatory diseases.
Keywords: Trex-1; cytokines; protein arginine deiminases; small-molecule inhibitor; stimulator of interferon genes.
Conflict of interest statement
K.A.F. serves on the scientific advisory board of NodThera Inc., Generation Bio, and Janssen and is a scientific founder of Danger Bio, LLC. P.R.T. is a scientific founder of Danger Bio. At the time of the study, S.-L.N. and G.S.P. were employed by GSK. All GSK authors have equity holdings in GSK. U.S. Patent Application No.: 63/147,387 Title: STING inhibitors filing Date: Feb 9, 2021 Inventor(s): F.H., L.S.-G., P.R.T., and K.A.F.
Figures


Similar articles
-
cGAS-mediated autophagy protects the liver from ischemia-reperfusion injury independently of STING.Am J Physiol Gastrointest Liver Physiol. 2018 Jun 1;314(6):G655-G667. doi: 10.1152/ajpgi.00326.2017. Epub 2018 Feb 15. Am J Physiol Gastrointest Liver Physiol. 2018. PMID: 29446653 Free PMC article.
-
STING-ΔN, a novel splice isoform of STING, modulates innate immunity and autophagy in response to DNA virus infection.Cell Commun Signal. 2025 Jun 21;23(1):299. doi: 10.1186/s12964-025-02305-w. Cell Commun Signal. 2025. PMID: 40544261 Free PMC article.
-
Stimulator of Interferon Genes (STING)-Type I Interferon Signaling: Bridging Immunity and Pain.J Integr Neurosci. 2025 Jun 23;24(6):33414. doi: 10.31083/JIN33414. J Integr Neurosci. 2025. PMID: 40613364 Review.
-
Dihydrotanshinone I, a main compound of Salvia miltiorrhiza, alleviates autoimmune and inflammatory diseases by directly targeting IRF3.Phytomedicine. 2025 Sep;145:157005. doi: 10.1016/j.phymed.2025.157005. Epub 2025 Jun 18. Phytomedicine. 2025. PMID: 40609382
-
Crosstalk between oxidative stress, mitochondrial dysfunction, chromosome instability, and the activation of the cGAS-STING/IFN pathway in systemic sclerosis.Ageing Res Rev. 2025 Aug;110:102812. doi: 10.1016/j.arr.2025.102812. Epub 2025 Jun 23. Ageing Res Rev. 2025. PMID: 40562314 Review.
Cited by
-
The balance of STING signaling orchestrates immunity in cancer.Nat Immunol. 2024 Jul;25(7):1144-1157. doi: 10.1038/s41590-024-01872-3. Epub 2024 Jun 25. Nat Immunol. 2024. PMID: 38918609 Review.
-
Development of nitroalkene-based inhibitors to target STING-dependent inflammation.Redox Biol. 2024 Aug;74:103202. doi: 10.1016/j.redox.2024.103202. Epub 2024 May 21. Redox Biol. 2024. PMID: 38865901 Free PMC article.
-
cGAS/STING signalling pathway in senescence and oncogenesis.Semin Cancer Biol. 2024 Nov;106-107:87-102. doi: 10.1016/j.semcancer.2024.08.007. Epub 2024 Aug 31. Semin Cancer Biol. 2024. PMID: 39222763 Review.
-
Applications of cryo-EM in drug development for STING.Curr Opin Struct Biol. 2024 Feb;84:102767. doi: 10.1016/j.sbi.2023.102767. Epub 2024 Jan 5. Curr Opin Struct Biol. 2024. PMID: 38183862 Free PMC article. Review.
-
The cGAS-STING pathway in cancer immunity: dual roles, therapeutic strategies, and clinical challenges.Essays Biochem. 2025 Mar 7;69(2):EBC20253006. doi: 10.1042/EBC20253006. Essays Biochem. 2025. PMID: 40052963 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

