Identification of scaffold proteins for improved endogenous engineering of extracellular vesicles
- PMID: 37550290
- PMCID: PMC10406850
- DOI: 10.1038/s41467-023-40453-0
Identification of scaffold proteins for improved endogenous engineering of extracellular vesicles
Abstract
Extracellular vesicles (EVs) are gaining ground as next-generation drug delivery modalities. Genetic fusion of the protein of interest to a scaffold protein with high EV-sorting ability represents a robust cargo loading strategy. To address the paucity of such scaffold proteins, we leverage a simple and reliable assay that can distinguish intravesicular cargo proteins from surface- as well as non-vesicular proteins and compare the EV-sorting potential of 244 candidate proteins. We identify 24 proteins with conserved EV-sorting abilities across five types of producer cells. TSPAN2 and TSPAN3 emerge as lead candidates and outperform the well-studied CD63 scaffold. Importantly, these engineered EVs show promise as delivery vehicles in cell cultures and mice as demonstrated by efficient transfer of luminal cargo proteins as well as surface display of different functional entities. The discovery of these scaffolds provides a platform for EV-based engineering.
© 2023. Springer Nature Limited.
Conflict of interest statement
A.G., D.G., J.Z.N. and S.E.A. are consultants for and have equity interests in EVOX Therapeutics Ltd, Oxford, UK. The other authors declare no competing interests.
Figures





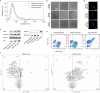
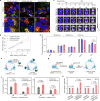
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

