Cas9-mediated knockout of Ndrg2 enhances the regenerative potential of dendritic cells for wound healing
- PMID: 37550295
- PMCID: PMC10406832
- DOI: 10.1038/s41467-023-40519-z
Cas9-mediated knockout of Ndrg2 enhances the regenerative potential of dendritic cells for wound healing
Abstract
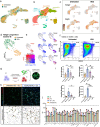
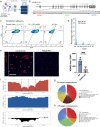
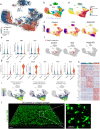
Chronic wounds impose a significant healthcare burden to a broad patient population. Cell-based therapies, while having shown benefits for the treatment of chronic wounds, have not yet achieved widespread adoption into clinical practice. We developed a CRISPR/Cas9 approach to precisely edit murine dendritic cells to enhance their therapeutic potential for healing chronic wounds. Using single-cell RNA sequencing of tolerogenic dendritic cells, we identified N-myc downregulated gene 2 (Ndrg2), which marks a specific population of dendritic cell progenitors, as a promising target for CRISPR knockout. Ndrg2-knockout alters the transcriptomic profile of dendritic cells and preserves an immature cell state with a strong pro-angiogenic and regenerative capacity. We then incorporated our CRISPR-based cell engineering within a therapeutic hydrogel for in vivo cell delivery and developed an effective translational approach for dendritic cell-based immunotherapy that accelerated healing of full-thickness wounds in both non-diabetic and diabetic mouse models. These findings could open the door to future clinical trials using safe gene editing in dendritic cells for treating various types of chronic wounds.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Nussbaum SR, et al. An economic evaluation of the impact, cost, and medicare policy implications of chronic nonhealing wounds. Value Health. 2018;21:27–32. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

