Asparagine restriction enhances CD8+ T cell metabolic fitness and antitumoral functionality through an NRF2-dependent stress response
- PMID: 37550596
- PMCID: PMC10447245
- DOI: 10.1038/s42255-023-00856-1
Asparagine restriction enhances CD8+ T cell metabolic fitness and antitumoral functionality through an NRF2-dependent stress response
Abstract
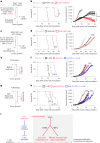
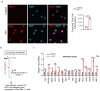
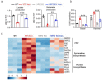
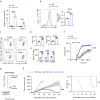
Robust and effective T cell immune surveillance and cancer immunotherapy require proper allocation of metabolic resources to sustain energetically costly processes, including growth and cytokine production. Here, we show that asparagine (Asn) restriction on CD8+ T cells exerted opposing effects during activation (early phase) and differentiation (late phase) following T cell activation. Asn restriction suppressed activation and cell cycle entry in the early phase while rapidly engaging the nuclear factor erythroid 2-related factor 2 (NRF2)-dependent stress response, conferring robust proliferation and effector function on CD8+ T cells during differentiation. Mechanistically, NRF2 activation in CD8+ T cells conferred by Asn restriction rewired the metabolic program by reducing the overall glucose and glutamine consumption but increasing intracellular nucleotides to promote proliferation. Accordingly, Asn restriction or NRF2 activation potentiated the T cell-mediated antitumoral response in preclinical animal models, suggesting that Asn restriction is a promising and clinically relevant strategy to enhance cancer immunotherapy. Our study revealed Asn as a critical metabolic node in directing the stress signaling to shape T cell metabolic fitness and effector functions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

