Long noncoding and micro-RNA expression in a model of articular chondrocyte degeneration induced by stromal cell-derived factor-1
- PMID: 37551168
- PMCID: PMC10321185
- DOI: 10.2478/abm-2022-0021
Long noncoding and micro-RNA expression in a model of articular chondrocyte degeneration induced by stromal cell-derived factor-1
Abstract
Background: Gene regulatory network analysis has found that long noncoding ribonucleic acids (lncRNAs) are strongly associated with the pathogenesis of osteoarthritis.
Objectives: To determine the differential expression of lncRNAs and microRNAs (miRNAs) in normal chondrocytes and those from a model of articular chondrocyte degeneration.
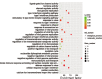
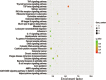
Methods: Chondrocytes were cultured from cartilage obtained from patients diagnosed with osteoarthritis of the knee. Stromal cell-derived factor-1 (SDF-1) was used to induce their degeneration. Total RNA was extracted, analyzed, amplified, labeled, and hybridized on a chip to determine expression. The set of enriched differentially expressed miRNAs was analyzed by gene ontology and the Kyoto Encyclopedia of Genes and Genomes to describe the functional properties of the key biological processes and pathways. We conducted a bioinformatics analysis using Cytoscape to elucidate the interactions between miRNAs and proteins.
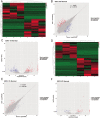
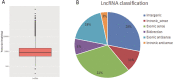
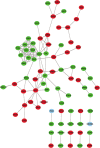
Results: We found that the expression of 186 lncRNAs was significantly different in the model of chondrocyte degeneration, in which 88 lncRNAs were upregulated, and 98 were downregulated. Expression of 684 miRNAs was significantly different. Analysis of the protein-protein interaction (PPI) network indicated that the genes for CXCL10, ISG15, MYC, MX1, OASL, IFIT1, RSAD2, MX2, IFI44L, and BST2 are the top 10 core genes, identifying the most important functional modules to elucidate the differential expression of miRNAs.
Conclusions: These data may provide new insights into the molecular mechanisms of chondrocyte degeneration in osteoarthritis, and the identification of lncRNAs and miRNAs may provide potential targets for the differential diagnosis and therapy of osteoarthritis.
Keywords: RNA; chondrocytes; high-throughput nucleotide sequencing; long noncoding; microRNAs; osteoarthritis.
© 2022 Guoliang Wang et al., published by Sciendo.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
