Evaluating the Frequencies of CNOT3, GRIA1, NFATC2, and PNPLA3 Variant Alleles and Their Association with L-Asparaginase Hypersensitivity in Pediatric Acute Lymphoblastic Leukemia in Addis Ababa, Ethiopia
- PMID: 37551203
- PMCID: PMC10404408
- DOI: 10.2147/TACG.S404695
Evaluating the Frequencies of CNOT3, GRIA1, NFATC2, and PNPLA3 Variant Alleles and Their Association with L-Asparaginase Hypersensitivity in Pediatric Acute Lymphoblastic Leukemia in Addis Ababa, Ethiopia
Abstract
Introduction: L-asparaginase is a vital component for the treatment of childhood acute lymphoblastic leukemia (ALL); however, hypersensitivity reactions and hepatotoxicity hinder its anti-neoplastic efficacy. Previous reports indicated that genetic variants in CNOT3, GRIA1, and NFATC2 genes might be associated with hypersensitivity reactions and PNPLA3 with liver function.
Objective: In this study, it was investigated whether this association also exists in a pediatric ALL cohort from Ethiopia.
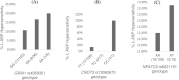
Methods: Three variants GRIA1 rs4958351, CNOT3 rs73062673, and NFATC2 rs6021191 were genotyped in a cohort of 160 patients. Association analysis to investigate the association with hypersensitivity reactions was performed using logistic regression analyses. Besides these variants, a variant in PNPLA3 (rs738409) was genotyped to assess the association with liver function.
Results: Genotype frequencies of GRIA1 rs4958351, CNOT3 rs73062673, and NFATC2 rs6021191 were higher/lower than previously reported. One hundred and forty-four patients were included in the association analysis of which, 18 (12.5%) developed L-ASP hypersensitivity. Though the frequency of hypersensitivity was higher in patients that carried the risk alleles of the three investigated genes, no statistically significant differences were observed. Association analysis between PNPLA3 rs738409 and liver function could not be investigated due to a lack of clinical information.
Conclusion: In conclusion, none of the tested genes did predict L-asparaginase hypersensitivity in an Ethiopian pediatric ALL patients.
Keywords: L-asparaginase; acute lymphoblastic leukemia; hypersensitivity.
© 2023 Ali et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
Similar articles
-
Impact of single-nucleotide variants and individual characteristics on adverse events of L-asparaginase in children with acute lymphoblastic leukemia.Front Pharmacol. 2024 Oct 24;15:1423049. doi: 10.3389/fphar.2024.1423049. eCollection 2024. Front Pharmacol. 2024. PMID: 39512835 Free PMC article.
-
Subgroups of Paediatric Acute Lymphoblastic Leukaemia Might Differ Significantly in Genetic Predisposition to Asparaginase Hypersensitivity.PLoS One. 2015 Oct 12;10(10):e0140136. doi: 10.1371/journal.pone.0140136. eCollection 2015. PLoS One. 2015. PMID: 26457809 Free PMC article.
-
HLA-DRB1*16:02 is associated with PEG-asparaginase hypersensitivity.Pharmacogenomics. 2021 Nov;22(17):1135-1142. doi: 10.2217/pgs-2021-0107. Epub 2021 Nov 8. Pharmacogenomics. 2021. PMID: 34747637
-
Review of pharmacogenetics studies of L-asparaginase hypersensitivity in acute lymphoblastic leukemia points to variants in the GRIA1 gene.Drug Metab Pers Ther. 2017 Mar 1;32(1):1-9. doi: 10.1515/dmpt-2016-0033. Drug Metab Pers Ther. 2017. PMID: 28259867 Review.
-
Insights into Asparaginase Allergic Responses: Exploring Pharmacogenetic Influences.Pharmaceutics. 2024 Aug 28;16(9):1134. doi: 10.3390/pharmaceutics16091134. Pharmaceutics. 2024. PMID: 39339172 Free PMC article. Review.
Cited by
-
Impact of single-nucleotide variants and individual characteristics on adverse events of L-asparaginase in children with acute lymphoblastic leukemia.Front Pharmacol. 2024 Oct 24;15:1423049. doi: 10.3389/fphar.2024.1423049. eCollection 2024. Front Pharmacol. 2024. PMID: 39512835 Free PMC article.
References
LinkOut - more resources
Full Text Sources