Deubiquitinase OTUD5 as a Novel Protector against 4-HNE-Triggered Ferroptosis in Myocardial Ischemia/Reperfusion Injury
- PMID: 37552043
- PMCID: PMC10558642
- DOI: 10.1002/advs.202301852
Deubiquitinase OTUD5 as a Novel Protector against 4-HNE-Triggered Ferroptosis in Myocardial Ischemia/Reperfusion Injury
Abstract
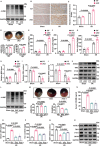
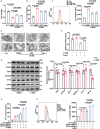
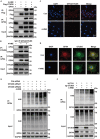
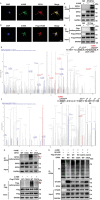
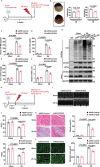
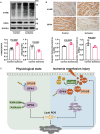
Despite the development of advanced technologies for interventional coronary reperfusion after myocardial infarction, a substantial number of patients experience high mortality due to myocardial ischemia-reperfusion (MI/R) injury. An in-depth understanding of the mechanisms underlying MI/R injury can provide crucial strategies for mitigating myocardial damage and improving patient survival. Here, it is discovered that the 4-hydroxy-2-nonenal (4-HNE) accumulates during MI/R, accompanied by high rates of myocardial ferroptosis. The loss-of-function of aldehyde dehydrogenase 2 (ALDH2), which dissipates 4-HNE, aggravates myocardial ferroptosis, whereas the activation of ALDH2 mitigates ferroptosis. Mechanistically, 4-HNE targets glutathione peroxidase 4 (GPX4) for K48-linked polyubiquitin-related degradation, which 4-HNE-GPX4 axis commits to myocyte ferroptosis and forms a positive feedback circuit. 4-HNE blocks the interaction between GPX4 and ovarian tumor (OTU) deubiquitinase 5 (OTUD5) by directly carbonylating their cysteine residues at C93 of GPX4 and C247 of OTUD5, identifying OTUD5 as the novel deubiquitinase for GPX4. Consequently, the elevation of OTUD5 deubiquitinates and stabilizes GPX4 to reverse 4-HNE-induced ferroptosis and alleviate MI/R injury. The data unravel the mechanism of 4-HNE in GPX4-dependent ferroptosis and identify OTUD5 as a novel therapeutic target for the treatment of MI/R injury.
Keywords: 4-hydroxy-2-nonenal (4-HNE); ferroptosis; glutathione peroxidase 4 (GPX4); myocardial ischemia reperfusion injury; ovarian tumor (OTU) deubiquitinase 5 (OTUD5).
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tsao C. W., Aday A. W., Almarzooq Z. I., Alonso A., Beaton A. Z., Bittencourt M. S., Boehme A. K., Buxton A. E., Carson A. P., Commodore‐Mensah Y., Elkind M. S. V., Evenson K. R., Eze‐Nliam C., Ferguson J. F., Generoso G., Ho J. E., Kalani R., Khan S. S., Kissela B. M., Knutson K. L., Levine D. A., Lewis T. T., Liu J., Loop M. S., Ma J., Mussolino M. E., Navaneethan S. D., Perak A. M., Poudel R., Rezk‐Hanna M., et al., Circulation 2022, 145, e153. - PubMed
-
- Ibáñez B., Heusch G., Ovize M., Van de Werf F., Journal of the American College of Cardiology 2015, 65, 1454. - PubMed
-
- Heusch G., Nat. Rev. Cardiol. 2020, 17, 773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82030059/State Key Program of the National Natural Science Foundation of China
- 82172178/National Natural Science Foundation of China
- 82072144/National Natural Science Foundation of China
- 81873950/National Natural Science Foundation of China
- 81873953/National Natural Science Foundation of China
- 81300219/National Natural Science Foundation of China
- 81671951/National Natural Science Foundation of China
- 2020YFC1512700/National Key R&D Program of China
- 2020YFC1512705/National Key R&D Program of China
- 2020YFC1512703/National Key R&D Program of China
- 2018FY100600/National S&T Fundamental Resources Investigation Project
- 2018FY100602/National S&T Fundamental Resources Investigation Project
- 2021ZLGX02/Key R&D Program of Shandong Province
- 2022ZLGX03/Key R&D Program of Shandong Province
- ZR2022MH078/Natural Science Foundation of Shandong Province
- 2019GSF108131/Key R&D Program of Shandong Province
- tsqn202103173/Taishan Young Scholar Program of Shandong Province
- tspd20181220/Taishan Pandeng Scholar Program of Shandong Province
- Youth Top-Talent Project of National Ten Thousand Talents Plan
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
